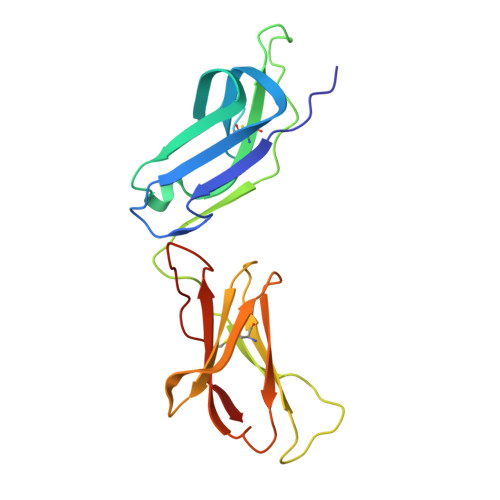
Structural basis for collagen recognition by the immune receptor OSCAR.
Zhou, L., Hinerman, J.M., Blaszczyk, M., Miller, J.L., Conrady, D.G., Barrow, A.D., Chirgadze, D.Y., Bihan, D., Farndale, R.W., Herr, A.B.(2016) Blood 127: 529-537
- PubMed: 26552697
- DOI: https://doi.org/10.1182/blood-2015-08-667055
- Primary Citation of Related Structures:
5EIQ, 5EIV - PubMed Abstract:
The osteoclast-associated receptor (OSCAR) is a collagen-binding immune receptor with important roles in dendritic cell maturation and activation of inflammatory monocytes as well as in osteoclastogenesis. The crystal structure of the OSCAR ectodomain is presented, both free and in complex with a consensus triple-helical peptide (THP). The structures revealed a collagen-binding site in each immunoglobulin-like domain (D1 and D2). The THP binds near a predicted collagen-binding groove in D1, but a more extensive interaction with D2 is facilitated by the unusually wide D1-D2 interdomain angle in OSCAR. Direct binding assays, combined with site-directed mutagenesis, confirm that the primary collagen-binding site in OSCAR resides in D2, in marked contrast to the related collagen receptors, glycoprotein VI (GPVI) and leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1). Monomeric OSCAR D1D2 binds to the consensus THP with a KD of 28 µM measured in solution, but shows a higher affinity (KD 1.5 μM) when binding to a solid-phase THP, most likely due to an avidity effect. These data suggest a 2-stage model for the interaction of OSCAR with a collagen fibril, with transient, low-affinity interactions initiated by the membrane-distal D1, followed by firm adhesion to the primary binding site in D2.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom;