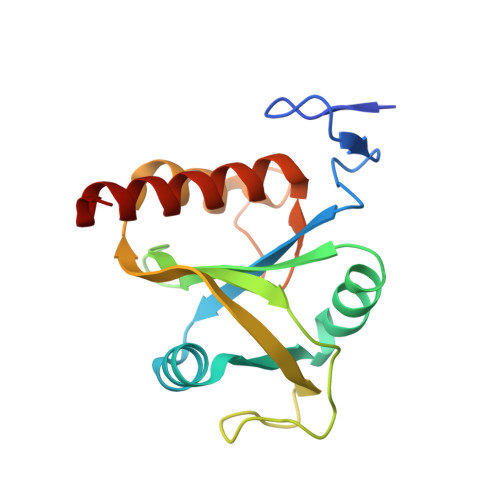
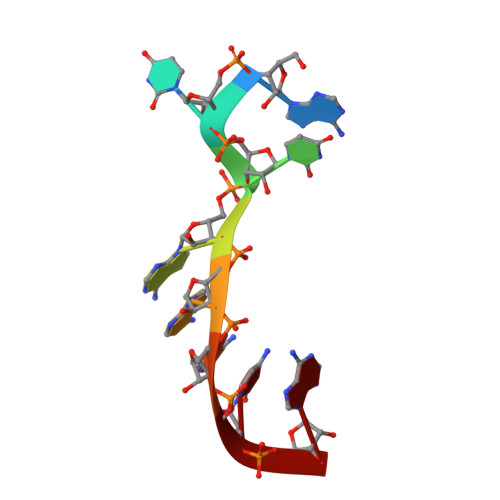Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1
Wu, B.X., Xu, J.H., Su, S.C., Liu, H., Gan, J., Ma, J.B.(2017) Biochem Biophys Res Commun 491: 310-316
- PubMed: 28735863
- DOI: https://doi.org/10.1016/j.bbrc.2017.07.104
- Primary Citation of Related Structures:
5EIM, 5EIP - PubMed Abstract:
Meiosis is one of the most dramatic differentiation programs accompanied by a striking change in gene expression profiles in fission yeast Schizosaccharomyces pombe. Whereas a number of meiosis-specific transcripts are expressed untimely in mitotic cells, and the entry of meiosis will be blocked as the accumulation of meiosis-specific mRNAs in the mitotic cells. A YTH domain containing protein Mmi1 was identified as a pivotal effector in a post-transcriptional event termed selective elimination of meiosis-specific mRNAs. Mmi1 can recognize and bind a class of meiosis-specific transcripts expressed inappropriately in mitotic cells, which all contain a conservative region called DSR, as a mark to remove them in cooperation with nuclear exosomes. Here we report the 1.6 Å resolution crystal structure of the Mmi1-YTH domain in complex with a high consensus hexanucleotide motif, which is multiple copied in the DSR region. Our structure observations, supported by site-directed mutations of key residues illustrate the mechanism for specific recognition of DSR-RNA by Mmi1. Moreover, different from other YTH domain family proteins, Mmi1-YTH domain has a distinctive RNA-binding properties although it has a similar fold as other ones.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Department of Biochemistry, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai, China. Electronic address: 15110700058@fudan.edu.cn.