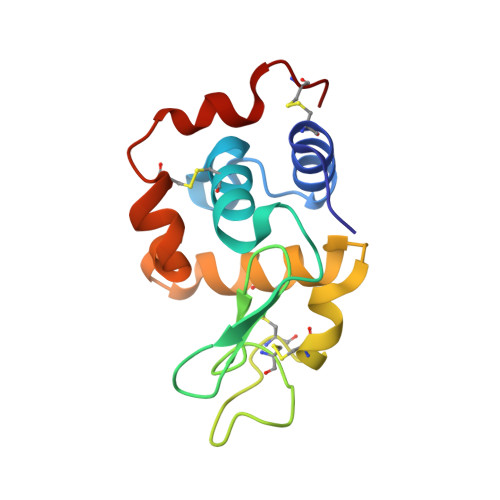
{Ru(CO)x}-core Complexes with N-Methylbenzimidazole (MBI) and 5,6-Dimethylbenzimidazole (DMBI) as Possible Metal Based Drugs and CO Release Materials. Synthesis, X-ray Structure and Evaluation of Cytotoxic Activity against Human Cancer Cells
Merlino, A.To be published.