Macromolecular diffractive imaging using imperfect crystals.
Ayyer, K., Yefanov, O.M., Oberthur, D., Roy-Chowdhury, S., Galli, L., Mariani, V., Basu, S., Coe, J., Conrad, C.E., Fromme, R., Schaffer, A., Dorner, K., James, D., Kupitz, C., Metz, M., Nelson, G., Xavier, P.L., Beyerlein, K.R., Schmidt, M., Sarrou, I., Spence, J.C., Weierstall, U., White, T.A., Yang, J.H., Zhao, Y., Liang, M., Aquila, A., Hunter, M.S., Robinson, J.S., Koglin, J.E., Boutet, S., Fromme, P., Barty, A., Chapman, H.N.(2016) Nature 530: 202-206
- PubMed: 26863980
- DOI: https://doi.org/10.1038/nature16949
- Primary Citation of Related Structures:
5E79, 5E7C - PubMed Abstract:
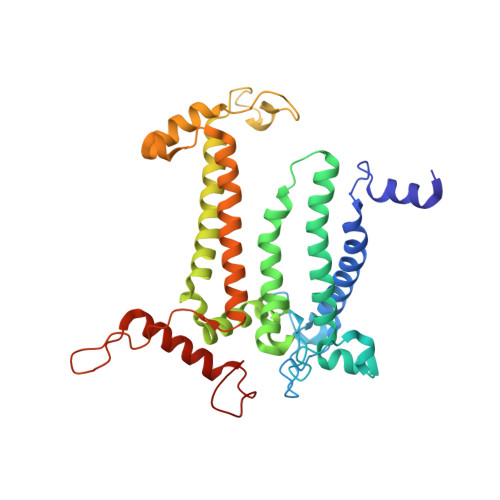
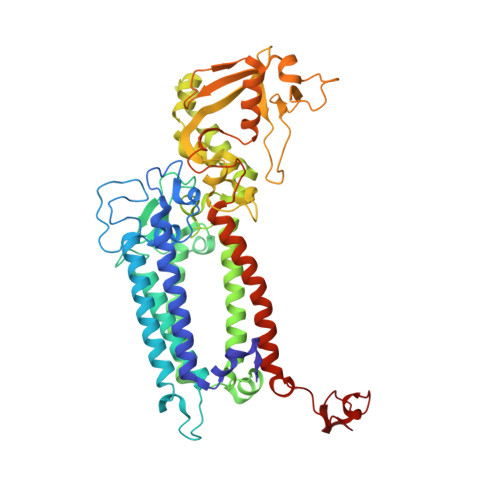
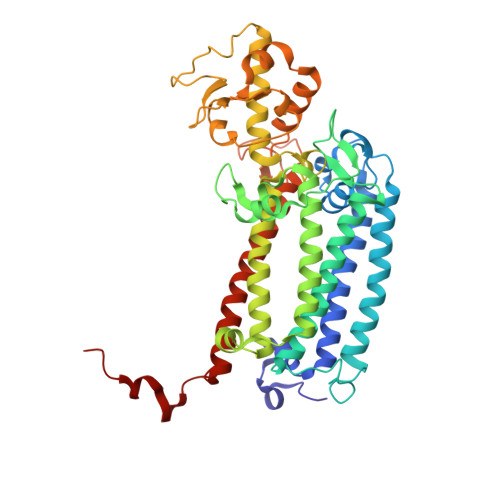
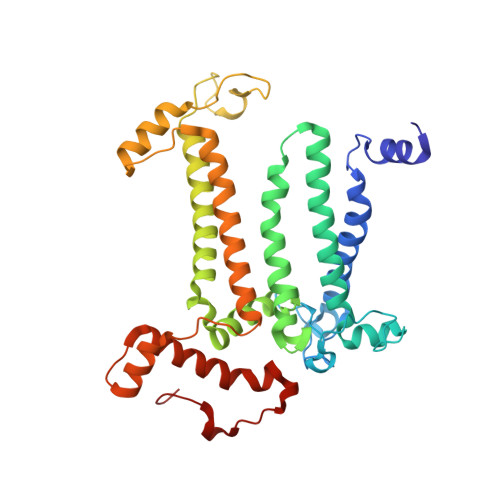
The three-dimensional structures of macromolecules and their complexes are mainly elucidated by X-ray protein crystallography. A major limitation of this method is access to high-quality crystals, which is necessary to ensure X-ray diffraction extends to sufficiently large scattering angles and hence yields information of sufficiently high resolution with which to solve the crystal structure. The observation that crystals with reduced unit-cell volumes and tighter macromolecular packing often produce higher-resolution Bragg peaks suggests that crystallographic resolution for some macromolecules may be limited not by their heterogeneity, but by a deviation of strict positional ordering of the crystalline lattice. Such displacements of molecules from the ideal lattice give rise to a continuous diffraction pattern that is equal to the incoherent sum of diffraction from rigid individual molecular complexes aligned along several discrete crystallographic orientations and that, consequently, contains more information than Bragg peaks alone. Although such continuous diffraction patterns have long been observed--and are of interest as a source of information about the dynamics of proteins--they have not been used for structure determination. Here we show for crystals of the integral membrane protein complex photosystem II that lattice disorder increases the information content and the resolution of the diffraction pattern well beyond the 4.5-ångström limit of measurable Bragg peaks, which allows us to phase the pattern directly. Using the molecular envelope conventionally determined at 4.5 ångströms as a constraint, we obtain a static image of the photosystem II dimer at a resolution of 3.5 ångströms. This result shows that continuous diffraction can be used to overcome what have long been supposed to be the resolution limits of macromolecular crystallography, using a method that exploits commonly encountered imperfect crystals and enables model-free phasing.
Organizational Affiliation:
Center for Free-Electron Laser Science, DESY, 22607 Hamburg, Germany.