Nuclear factor 90 uses an ADAR2-like binding mode to recognize specific bases in dsRNA.
Jayachandran, U., Grey, H., Cook, A.G.(2016) Nucleic Acids Res 44: 1924-1936
- PubMed: 26712564
- DOI: https://doi.org/10.1093/nar/gkv1508
- Primary Citation of Related Structures:
5DV7 - PubMed Abstract:
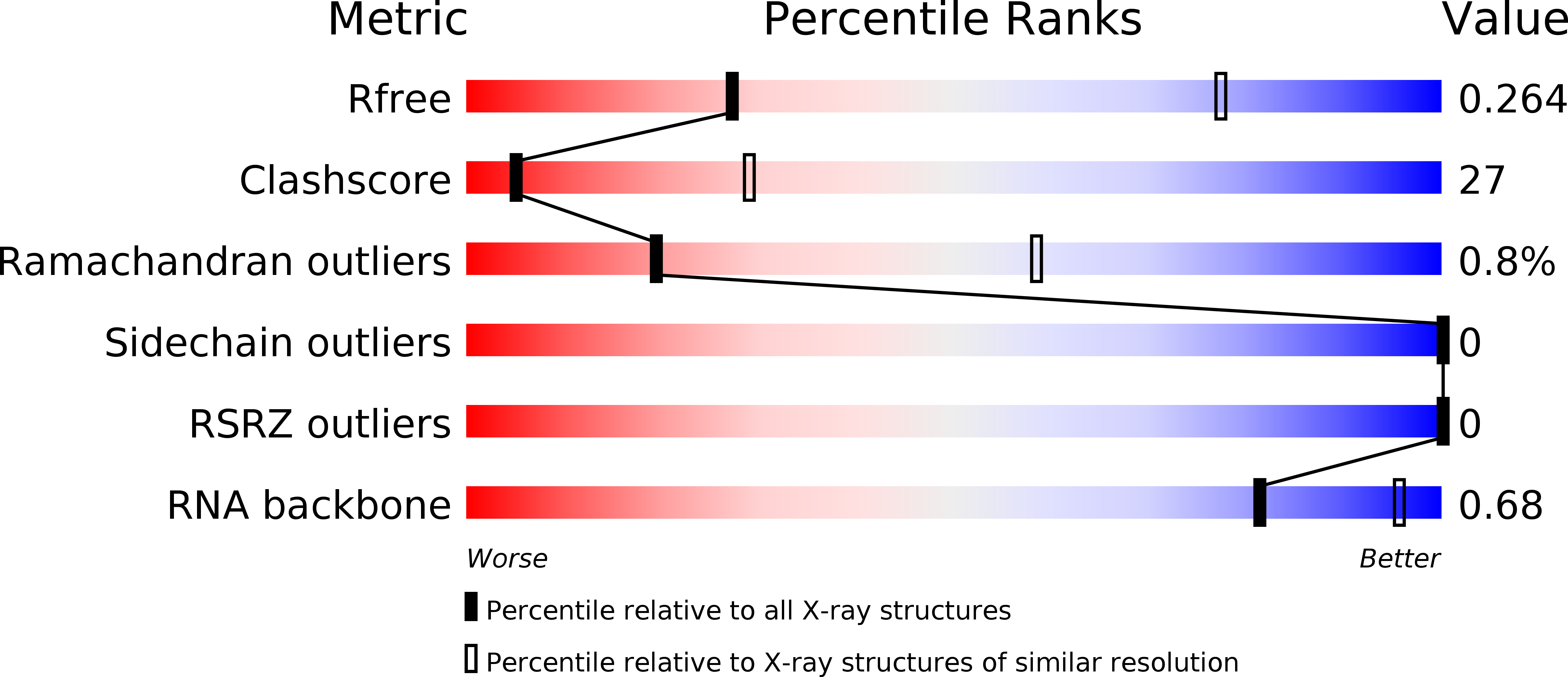
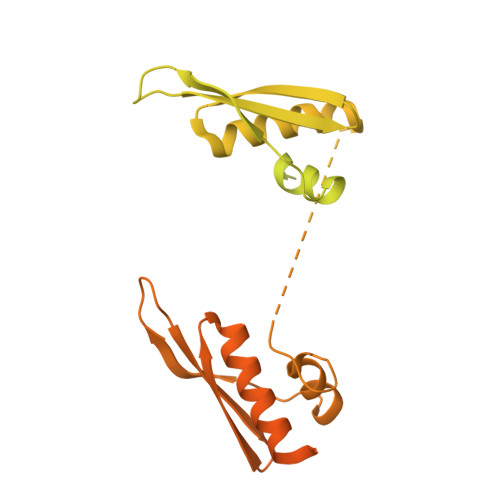
Nuclear factors 90 and 45 (NF90 and NF45) form a protein complex involved in the post-transcriptional control of many genes in vertebrates. NF90 is a member of the dsRNA binding domain (dsRBD) family of proteins. RNA binding partners identified so far include elements in 3' untranslated regions of specific mRNAs and several non-coding RNAs. In NF90, a tandem pair of dsRBDs separated by a natively unstructured segment confers dsRNA binding activity. We determined a crystal structure of the tandem dsRBDs of NF90 in complex with a synthetic dsRNA. This complex shows surprising similarity to the tandem dsRBDs from an adenosine-to-inosine editing enzyme, ADAR2 in complex with a substrate RNA. Residues involved in unusual base-specific recognition in the minor groove of dsRNA are conserved between NF90 and ADAR2. These data suggest that, like ADAR2, underlying sequences in dsRNA may influence how NF90 recognizes its target RNAs.
Organizational Affiliation:
Wellcome Trust Centre for Cell Biology, University of Edinburgh, Michael Swann Building, Max Born Crescent, Edinburgh EH9 3BF, UK.