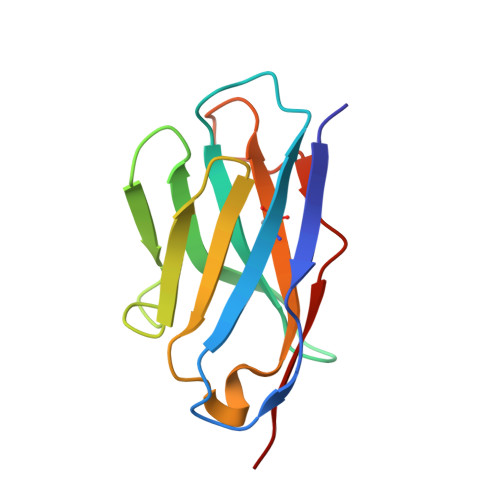
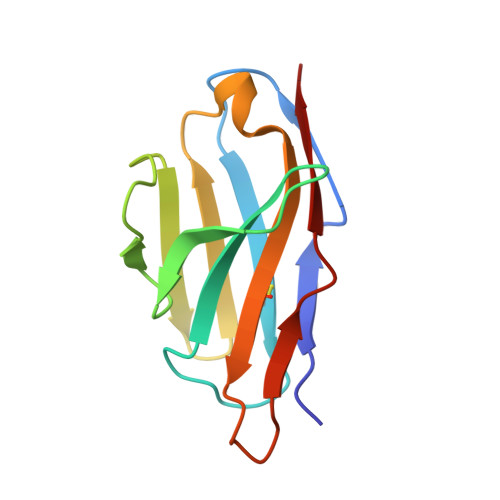
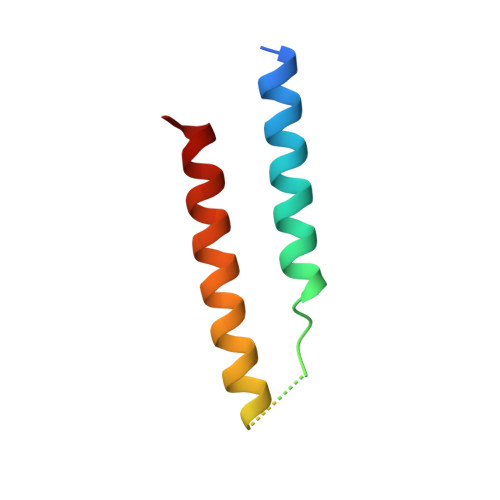
The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
DiMattia, M.A., Watts, N.R., Cheng, N., Huang, R., Heymann, J.B., Grimes, J.M., Wingfield, P.T., Stuart, D.I., Steven, A.C.(2016) Structure 24: 1068-1080
- PubMed: 27265851
- DOI: https://doi.org/10.1016/j.str.2016.04.015
- Primary Citation of Related Structures:
5DHV, 5DHX, 5DHY, 5DHZ - PubMed Abstract:
HIV-1 Rev protein mediates the nuclear export of viral RNA genomes. To do so, Rev oligomerizes cooperatively onto an RNA motif, the Rev response element (RRE), forming a complex that engages with the host nuclear export machinery. To better understand Rev oligomerization, we determined four crystal structures of Rev N-terminal domain dimers, which show that they can pivot about their dyad axis, giving crossing angles of 90° to 140°. In parallel, we performed cryoelectron microscopy of helical Rev filaments. Filaments vary from 11 to 15 nm in width, reflecting variations in dimer crossing angle. These structures contain additional density, indicating that C-terminal domains become partially ordered in the context of filaments. This conformational variability may be exploited in the assembly of RRE/Rev complexes. Our data also revealed a third interface between Revs, which offers an explanation for how the arrangement of Rev subunits adapts to the "A"-shaped architecture of the RRE in export-active complexes.
Organizational Affiliation:
Laboratory of Structural Biology Research, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892, USA; Division of Structural Biology, Henry Wellcome Building for Genomic Medicine, University of Oxford, Roosevelt Drive, Headington OX3 7BN, UK.