The crystal structure of methionine gamma-lyase from Citrobacter freundii, S339A mutant
Revtovich, S.V., Nikulin, A.D., Anufrieva, N.V., Morozova, E.A., Demidkina, T.V.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methionine gamma-lyase | 398 | Citrobacter freundii | Mutation(s): 1 Gene Names: TN42_08855, TO64_18625 EC: 4.4.1.11 | 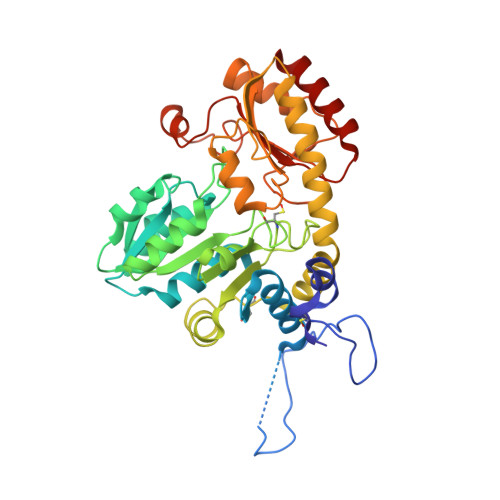 | |
UniProt | |||||
Find proteins for A0A0A5P8W7 (Citrobacter freundii) Explore A0A0A5P8W7 Go to UniProtKB: A0A0A5P8W7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0A5P8W7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PLP Query on PLP | D [auth A] | PYRIDOXAL-5'-PHOSPHATE C8 H10 N O6 P NGVDGCNFYWLIFO-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | B [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| NA Query on NA | C [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSO Query on CSO | A | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 56.659 | α = 90 |
| b = 123.09 | β = 90 |
| c = 128.785 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MxCuBE | data collection |
| XDS | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| RFBR | Russian Federation | -- |