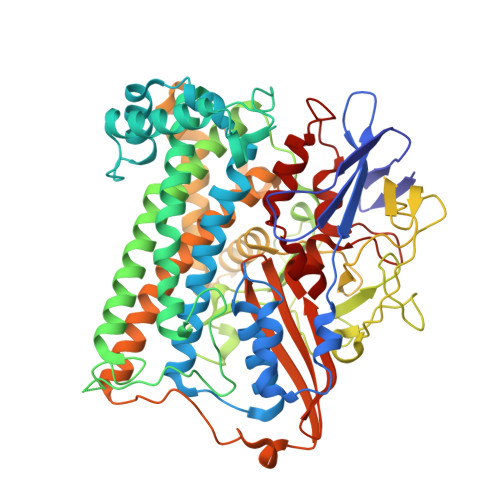
Krypton Derivatization of an O2 -Tolerant Membrane-Bound [NiFe] Hydrogenase Reveals a Hydrophobic Tunnel Network for Gas Transport.
Kalms, J., Schmidt, A., Frielingsdorf, S., van der Linden, P., von Stetten, D., Lenz, O., Carpentier, P., Scheerer, P.(2016) Angew Chem Int Ed Engl 55: 5586-5590
- PubMed: 26913499
- DOI: https://doi.org/10.1002/anie.201508976
- Primary Citation of Related Structures:
5D51 - PubMed Abstract:
[NiFe] hydrogenases are metalloenzymes catalyzing the reversible heterolytic cleavage of hydrogen into protons and electrons. Gas tunnels make the deeply buried active site accessible to substrates and inhibitors. Understanding the architecture and function of the tunnels is pivotal to modulating the feature of O2 tolerance in a subgroup of these [NiFe] hydrogenases, as they are interesting for developments in renewable energy technologies. Here we describe the crystal structure of the O2 -tolerant membrane-bound [NiFe] hydrogenase of Ralstonia eutropha (ReMBH), using krypton-pressurized crystals. The positions of the krypton atoms allow a comprehensive description of the tunnel network within the enzyme. A detailed overview of tunnel sizes, lengths, and routes is presented from tunnel calculations. A comparison of the ReMBH tunnel characteristics with crystal structures of other O2 -tolerant and O2 -sensitive [NiFe] hydrogenases revealed considerable differences in tunnel size and quantity between the two groups, which might be related to the striking feature of O2 tolerance.
Organizational Affiliation:
Institut für Medizinische Physik und Biophysik (CC2), Group Protein X-ray Crystallography and Signal Transduction, Charité - Universitätsmedizin Berlin, Charitéplatz 1, 10117, Berlin, Germany.