Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
Hirai, H., Yasui, N., Yamashita, K., Tabata, S., Yamamoto, M., Takagi, J., Nogi, T.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reelin | 725 | Mus musculus | Mutation(s): 1 Gene Names: Reln, Rl EC: 3.4.21 | 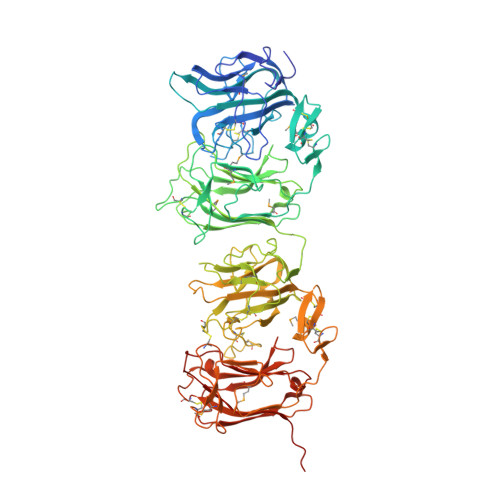 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q60841 (Mus musculus) Explore Q60841 Go to UniProtKB: Q60841 | |||||
IMPC: MGI:103022 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q60841 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, isoform CRA_e | 570 | Homo sapiens | Mutation(s): 0 Gene Names: LRP8, hCG_33395 | 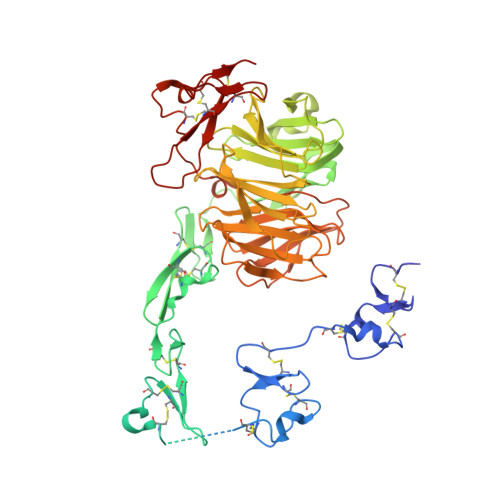 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q14114 (Homo sapiens) Explore Q14114 Go to UniProtKB: Q14114 | |||||
PHAROS: Q14114 GTEx: ENSG00000157193 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q14114 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | I [auth A], J [auth A], O [auth B], T [auth C], Y [auth D] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| CA Query on CA | E [auth A] F [auth A] G [auth A] H [auth A] K [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, C | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 205.951 | α = 90 |
| b = 205.951 | β = 90 |
| c = 169.84 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| autoSHARP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| JSPS | Japan | 22247010 |
| JSPS | Japan | 20770084 |
| JSPS | Japan | 05J09821 |
| MEXT | Japan | 17082004 |