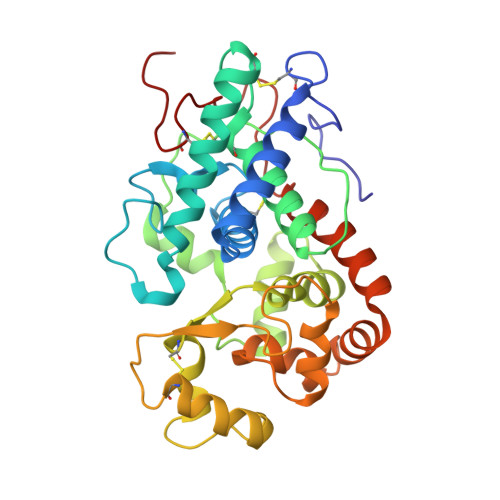
Structural and Spectroscopic Characterisation of a Heme Peroxidase from Sorghum.
Nnamchi, C.I., Parkin, G., Efimov, I., Basran, J., Kwon, H., Svistunenko, D.A., Agirre, J., Okolo, B.N., Moneke, A., Nwanguma, B.C., Moody, P.C.E., Raven, E.L.(2016) J Biol Inorg Chem 21: 63
- PubMed: 26666777
- DOI: https://doi.org/10.1007/s00775-015-1313-z
- Primary Citation of Related Structures:
5AOG - PubMed Abstract:
A cationic class III peroxidase from Sorghum bicolor was purified to homogeneity. The enzyme contains a high-spin heme, as evidenced by UV-visible spectroscopy and EPR. Steady state oxidation of guaiacol was demonstrated and the enzyme was shown to have higher activity in the presence of calcium ions. A Fe(III)/Fe(II) reduction potential of -266 mV vs NHE was determined. Stopped-flow experiments with H2O2 showed formation of a typical peroxidase Compound I species, which converts to Compound II in the presence of calcium. A crystal structure of the enzyme is reported, the first for a sorghum peroxidase. The structure reveals an active site that is analogous to those for other class I heme peroxidase, and a substrate binding site (assigned as arising from binding of indole-3-acetic acid) at the γ-heme edge. Metal binding sites are observed in the structure on the distal (assigned as a Na(+) ion) and proximal (assigned as a Ca(2+)) sides of the heme, which is consistent with the Ca(2+)-dependence of the steady state and pre-steady state kinetics. It is probably the case that the structural integrity (and, thus, the catalytic activity) of the sorghum enzyme is dependent on metal ion incorporation at these positions.
Organizational Affiliation:
Department of Microbiology, University of Nigeria, Nsukka, Nigeria.