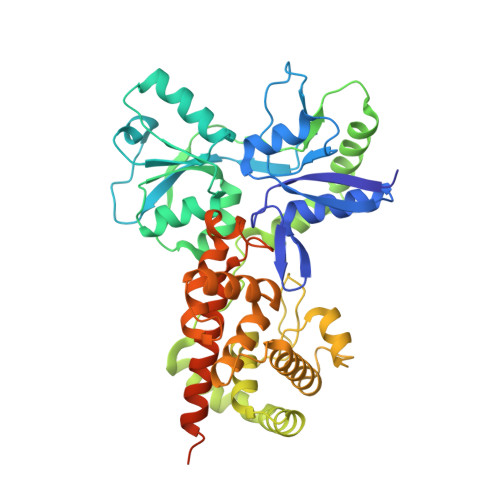
Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Dominguez-Gil, T., Lee, M., Acebron-Avalos, I., Mahasenan, K.V., Hesek, D., Dik, D.A., Byun, B., Lastochkin, E., Fisher, J.F., Mobashery, S., Hermoso, J.A.(2016) Structure 24: 1729-1741
- PubMed: 27618662
- DOI: https://doi.org/10.1016/j.str.2016.07.019
- Primary Citation of Related Structures:
5A5X, 5AA1, 5AA2, 5AA3, 5AA4 - PubMed Abstract:
Bacteria grow and divide without loss of cellular integrity. This accomplishment is notable, as a key component of their cell envelope is a surrounding glycopeptide polymer. In Gram-negative bacteria this polymer-the peptidoglycan-grows by the difference between concurrent synthesis and degradation. The regulation of the enzymatic ensemble for these activities is poorly understood. We report herein the structural basis for the control of one such enzyme, the lytic transglycosylase MltF of Pseudomonas aeruginosa. Its structure comprises two modules: an ABC-transporter-like regulatory module and a catalytic module. Occupancy of the regulatory module by peptidoglycan-derived muropeptides effects a dramatic and long-distance (40 Å) conformational change, occurring over the entire protein structure, to open its active site for catalysis. This discovery of the molecular basis for the allosteric control of MltF catalysis is foundational to further study of MltF within the complex enzymatic orchestration of the dynamic peptidoglycan.
Organizational Affiliation:
Departamento de Cristalografía y Biología Estructural, Instituto de Química-Física "Rocasolano", CSIC, Serrano 119, 28006 Madrid, Spain.