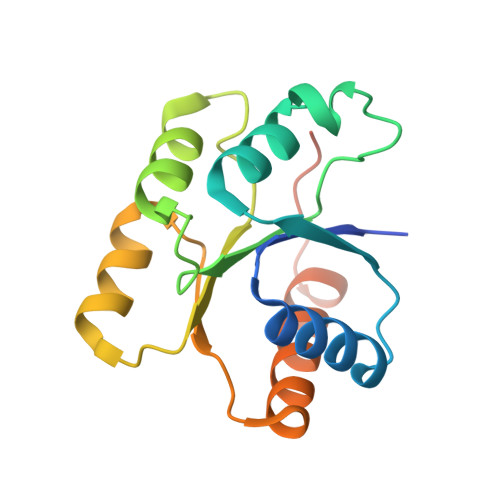
Structure of the response regulator RPA3017 involved in red-light signaling in Rhodopseudomonas palustris.
Yang, X., Zeng, X., Moffat, K., Yang, X.(2015) Acta Crystallogr F Struct Biol Commun 71: 1215-1222
- PubMed: 26457509
- DOI: https://doi.org/10.1107/S2053230X15014661
- Primary Citation of Related Structures:
4ZYL - PubMed Abstract:
Two-component signal transduction is the major signaling mechanism that enables bacteria to survive and thrive in complex environmental conditions. The photosynthetic bacterium R. palustris employs two tandem bacteriophytochromes, RpBphP2 and RpBphP3, to perceive red-light signals that regulate the synthesis of light-harvesting complexes under low-light conditions. Both RpBphP2 and RpBphP3 are photosensory histidine kinases coupled to the same response regulator RPA3017. Together, they constitute a two-component system that converts a red-light signal into a biological signal. In this work, the crystal structure of RPA3017 in the unphosphorylated form at 1.9 Å resolution is presented. This structure reveals a tightly associated dimer arrangement that is conserved among phytochrome-related response regulators. The conserved active-site architecture provides structural insight into the phosphotransfer reaction between RpBphP2/RpBphP3 and RPA3017. Based on structural comparisons and homology modeling, how specific recognition between RpBphP2/RpBphP3 and RPA3017 is achieved at the molecular level is further explored.
Organizational Affiliation:
National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, People's Republic of China.