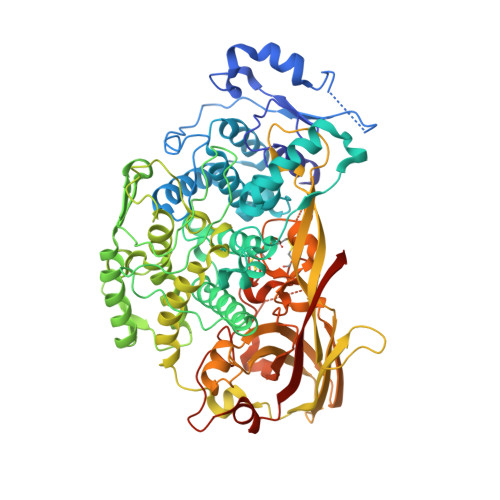
The structure of a prophenoloxidase (PPO) from Anopheles gambiae provides new insights into the mechanism of PPO activation.
Hu, Y., Wang, Y., Deng, J., Jiang, H.(2016) BMC Biol 14: 2-2
- PubMed: 26732497
- DOI: https://doi.org/10.1186/s12915-015-0225-2
- Primary Citation of Related Structures:
4YZW - PubMed Abstract:
Phenoloxidase (PO)-catalyzed melanization is a universal defense mechanism of insects against pathogenic and parasitic infections. In mosquitos such as Anopheles gambiae, melanotic encapsulation is a resistance mechanism against certain parasites that cause malaria and filariasis. PO is initially synthesized by hemocytes and released into hemolymph as inactive prophenoloxidase (PPO), which is activated by a serine protease cascade upon recognition of foreign invaders. The mechanisms of PPO activation and PO catalysis have been elusive.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, OK, 74078, USA. mianmian.hu@okstate.edu.