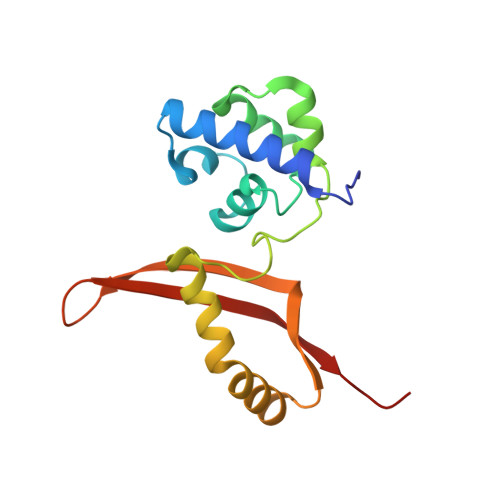
Serendipitous crystallization and structure determination of cyanase (CynS) from Serratia proteamaculans
Butryn, A., Stoehr, G., Linke-Winnebeck, C., Hopfner, K.-P.(2015) Acta Crystallogr F Struct Biol Commun 71: 471-476
- PubMed: 25849512
- DOI: https://doi.org/10.1107/S2053230X15004902
- Primary Citation of Related Structures:
4Y42 - PubMed Abstract:
Cyanate hydratase (CynS) catalyzes the decomposition of cyanate and bicarbonate into ammonia and carbon dioxide. Here, the serendipitous crystallization of CynS from Serratia proteamaculans (SpCynS) is reported. SpCynS was crystallized as an impurity and its identity was determined using mass-spectrometric analysis. The crystals belonged to space group P1 and diffracted to 2.1 Å resolution. The overall structure of SpCynS is very similar to a previously determined structure of CynS from Escherichia coli. Density for a ligand bound to the SpCynS active site was observed, but could not be unambiguously identified. Additionally, glycerol molecules bound at the entry to the active site of the enzyme indicate conserved residues that might be important for the trafficking of substrates and products.
Organizational Affiliation:
Gene Center and Department of Biochemistry, Ludwig-Maximilians-University Munich, Feodor-Lynen-Strasse 25, 81377 Munich, Germany.