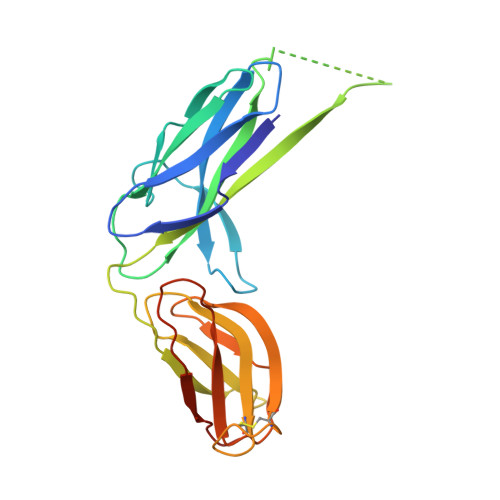
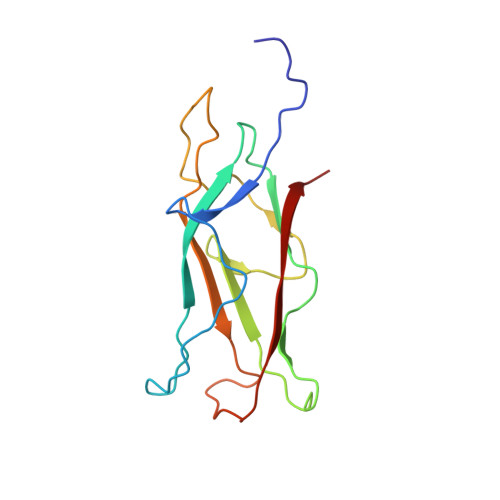
Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Bao, R., Liu, Y., Savarino, S.J., Xia, D.(2016) Mol Microbiol 102: 975-991
- PubMed: 27627030
- DOI: https://doi.org/10.1111/mmi.13530
- Primary Citation of Related Structures:
4Y2L, 4Y2N, 4Y2O - PubMed Abstract:
The assembly of the class 5 colonization factor antigen I (CFA/I) fimbriae of enterotoxigenic E. coli was proposed to proceed via the alternate chaperone-usher pathway. Here, we show that in the absence of the chaperone CfaA, CfaB, the major pilin subunit of CFA/I fimbriae, is able to spontaneously refold and polymerize into cyclic trimers. CfaA kinetically traps CfaB to form a metastable complex that can be stabilized by mutations. Crystal structure of the stabilized complex reveals distinctive interactions provided by CfaA to trap CfaB in an assembly competent state through donor-strand complementation (DSC) and cleft-mediated anchorage. Mutagenesis indicated that DSC controls the stability of the chaperone-subunit complex and the cleft-mediated anchorage of the subunit C-terminus additionally assist in subunit refolding. Surprisingly, over-stabilization of the chaperone-subunit complex led to delayed fimbria assembly, whereas destabilizing the complex resulted in no fimbriation. Thus, CfaA acts predominantly as a kinetic trap by stabilizing subunit to avoid its off-pathway self-polymerization that results in energetically favorable trimers and could serve as a driving force for CFA/I pilus assembly, representing an energetic landscape unique to class 5 fimbria assembly.
Organizational Affiliation:
Division of Infectious Diseases, National Key Laboratory of Biotherapy/Collaborative Innovation Center of Biotherapy, West China Hospitals, Sichuan University, Chengdu, 610041, China.