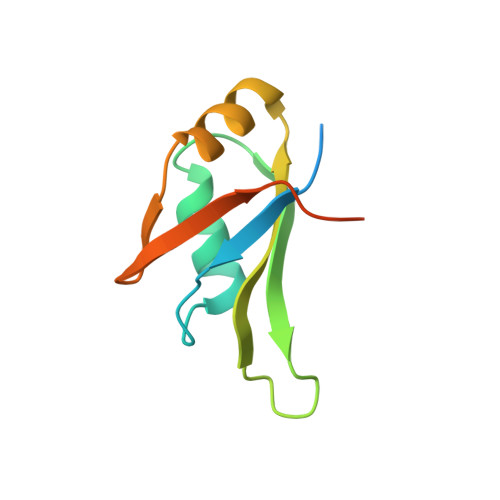
Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Chiang, C.H., Grauffel, C., Wu, L.S., Kuo, P.H., Doudeva, L.G., Lim, C., Shen, C.K., Yuan, H.S.(2016) Sci Rep 6: 21581-21581
- PubMed: 26883171
- DOI: https://doi.org/10.1038/srep21581
- Primary Citation of Related Structures:
4Y00, 4Y0F - PubMed Abstract:
The RNA-binding protein TDP-43 forms intracellular inclusions in amyotrophic lateral sclerosis (ALS). While TDP-43 mutations have been identified in ALS patients, how these mutations are linked to ALS remains unclear. Here we examined the biophysical properties of six ALS-linked TDP-43 mutants and found that one of the mutants, D169G, had higher thermal stability than wild-type TDP-43 and that it was cleaved by caspase 3 more efficiently, producing increased levels of the C-terminal 35 kD fragments (TDP-35) in vitro and in neuroblastoma cells. The crystal structure of the TDP-43 RRM1 domain containing the D169G mutation in complex with DNA along with molecular dynamics simulations reveal that the D169G mutation induces a local conformational change in a β turn and increases the hydrophobic interactions in the RRM1 core, thus enhancing the thermal stability of the RRM1 domain. Our results provide the first crystal structure of TDP-43 containing a disease-linked D169G mutation and a disease-related mechanism showing that D169G mutant is more susceptible to proteolytic cleavage by caspase 3 into the pathogenic C-terminal 35-kD fragments due to its increased stability in the RRM1 domain. Modulation of TDP-43 stability and caspase cleavage efficiency could present an avenue for prevention and treatment of TDP-43-linked neurodegeneration.
Organizational Affiliation:
Institute of Molecular Biology, Academia Sinica, Taipei, 11529, Taiwan.