Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Jang, T., Kim, S.H., Jeong, J., Kim, S., Kim, Y.G., Park, H.H.(2015) Sci Rep 5: 9847
- PubMed: 26038885
- DOI: https://doi.org/10.1038/srep09847
- Primary Citation of Related Structures:
4UZ0 - PubMed Abstract:
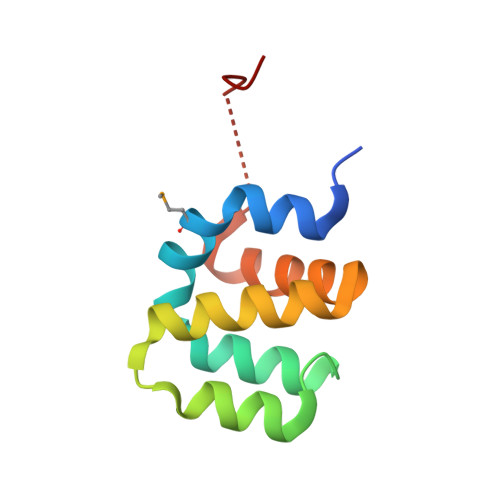
Apoptosis repressor with caspase recruiting domain (ARC) is a multifunctional inhibitor of apoptosis that is unusually over-expressed or activated in various cancers and in the state of the pulmonary hypertension. Therefore, ARC might be an optimal target for therapeutic intervention. Human ARC is composed of two distinct domains, N-terminal caspase recruiting domain (CARD) and C-terminal P/E (proline and glutamic acid) rich domain. ARC inhibits the extrinsic apoptosis pathway by interfering with DISC formation. ARC CARD directly interacts with the death domains (DDs) of Fas and FADD, as well as with the death effector domains (DEDs) of procaspase-8. Here, we report the first crystal structure of the CARD domain of ARC at a resolution of 2.4 Å. Our structure was a dimer with novel homo-dimerization interfaces that might be critical to its inhibitory function. Interestingly, ARC did not exhibit a typical death domain fold. The sixth helix (H6), which was detected at the typical death domain fold, was not detected in the structure of ARC, indicating that H6 may be dispensable for the function of the death domain superfamily.
Organizational Affiliation:
Department of Biochemistry, Yeungnam University, Gyeongsan, 712-749, South Korea.