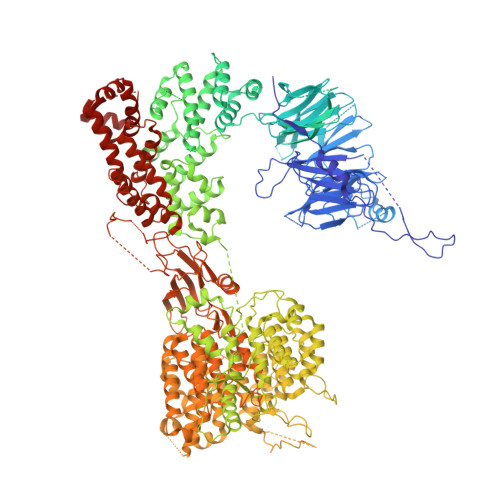
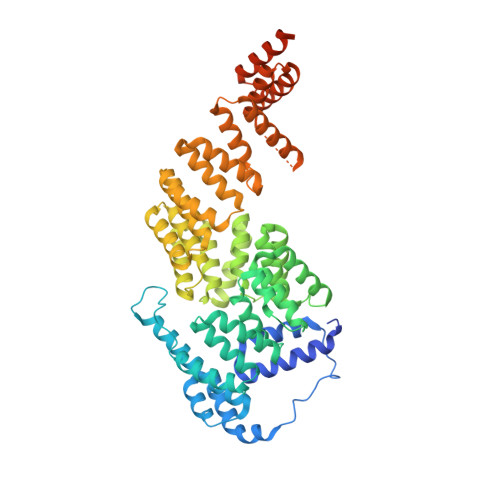
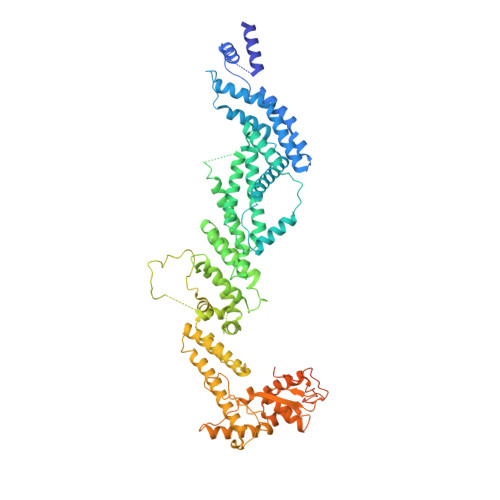
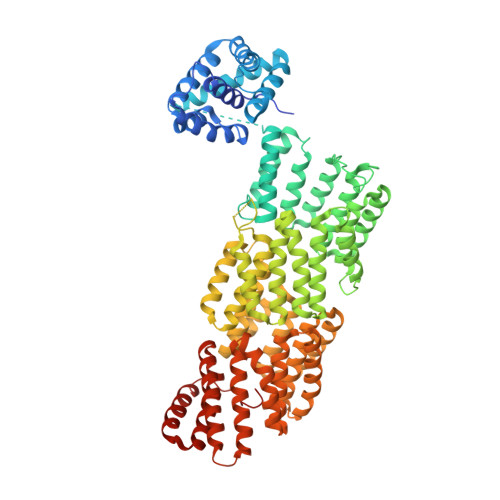
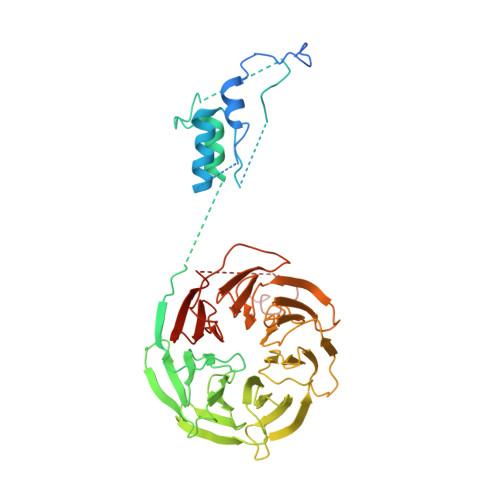
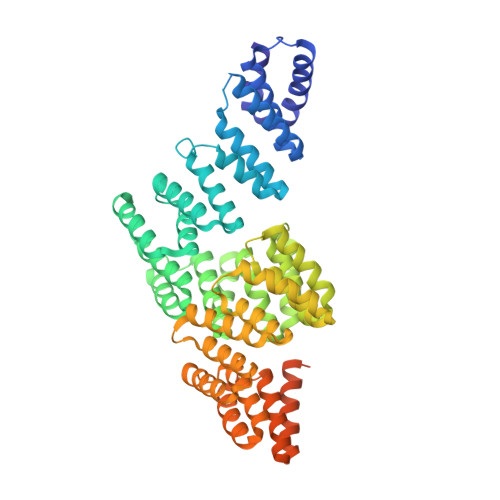
Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Chang, L., Zhang, Z., Yang, J., Mclaughlin, S.H., Barford, D.(2015) Nature 522: 450
- PubMed: 26083744
- DOI: https://doi.org/10.1038/nature14471
- Primary Citation of Related Structures:
4UI9, 5A31 - PubMed Abstract:
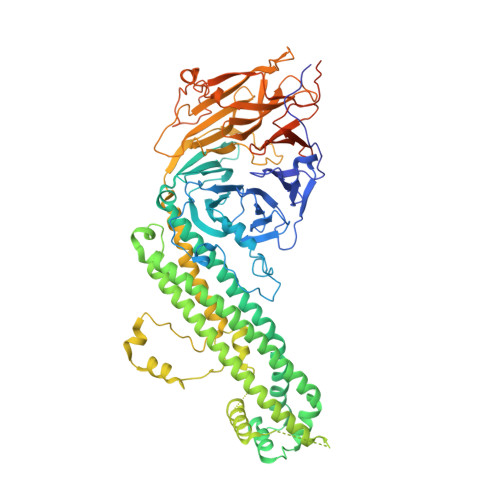
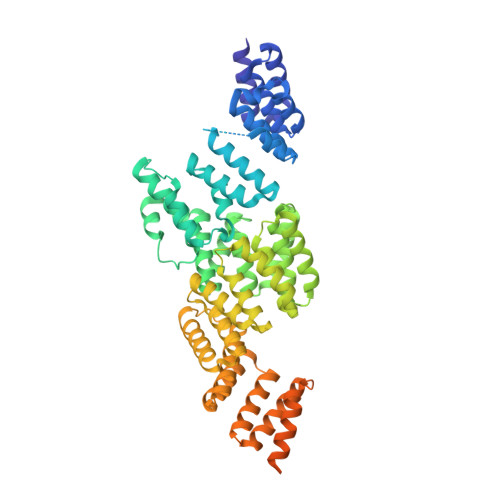
The anaphase-promoting complex (APC/C) is a multimeric RING E3 ubiquitin ligase that controls chromosome segregation and mitotic exit. Its regulation by coactivator subunits, phosphorylation, the mitotic checkpoint complex and interphase early mitotic inhibitor 1 (Emi1) ensures the correct order and timing of distinct cell-cycle transitions. Here we use cryo-electron microscopy to determine atomic structures of APC/C-coactivator complexes with either Emi1 or a UbcH10-ubiquitin conjugate. These structures define the architecture of all APC/C subunits, the position of the catalytic module and explain how Emi1 mediates inhibition of the two E2s UbcH10 and Ube2S. Definition of Cdh1 interactions with the APC/C indicates how they are antagonized by Cdh1 phosphorylation. The structure of the APC/C with UbcH10-ubiquitin reveals insights into the initiating ubiquitination reaction. Our results provide a quantitative framework for the design of future experiments to investigate APC/C functions in vivo.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge, CB2 0QH, UK.