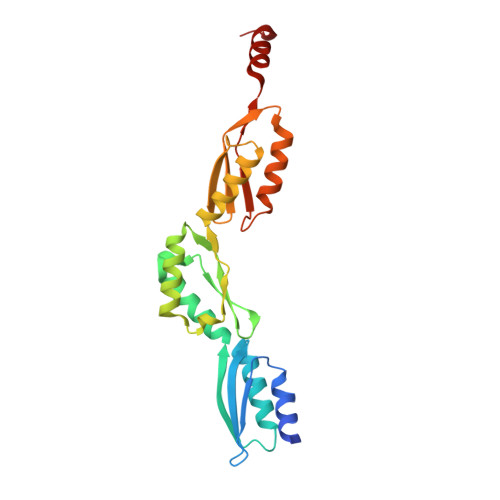
The extended structure of the periplasmic region of CdsD, a structural protein of the type III secretion system of Chlamydia trachomatis.
Merilainen, G., Koski, M.K., Wierenga, R.K.(2016) Protein Sci 25: 987-998
- PubMed: 26914207
- DOI: https://doi.org/10.1002/pro.2906
- Primary Citation of Related Structures:
4QO6, 4QQ0 - PubMed Abstract:
The type III secretion system (T3SS) is required for the virulence of many gram-negative bacterial human pathogens. It is composed of several structural proteins, forming the secretion needle and its basis, the basal body. In Chlamydia spp., the T3SS inner membrane ring (IM-ring) of the basal body is formed by the periplasmic part of CdsD (outer ring) and CdsJ (inner ring). Here we describe the crystal structure of the C-terminal, periplasmic part of CdsD, not including the last 60 residues. Two crystal forms were obtained, grown in three different crystallization conditions. In both crystal forms there is one molecule per asymmetric unit adopting a similar extended structure. The structures consist of three periplasmic domains (PDs) of similar αββαβ topology as seen also in the structures of the homologous PrgH (Salmonella typhimurium) and YscD (Yersinia enterocolitica). Only in the C2 crystal form, there is a C-terminal additional helix after the PD3 domain. The relative orientation of the three subsequent CdsD PD domains with respect to each other is more extended than in PrgH but less extended than in YscD. Small-angle X-ray scattering data show that also in solution this CdsD construct adopts the same elongated shape. In both crystal forms the CdsD molecules are packed in a parallel fashion, using translational crystallographic symmetry. The most extensive crystal contacts are preserved in both crystal forms, suggesting a possible mode of assembly of the CdsD periplasmic part into a 24-mer complex forming the outer ring of the IM-ring of the T3SS.
Organizational Affiliation:
Biocenter Oulu and Faculty of Biochemistry and Molecular Medicine, 90014 University of Oulu, Oulu, Finland.