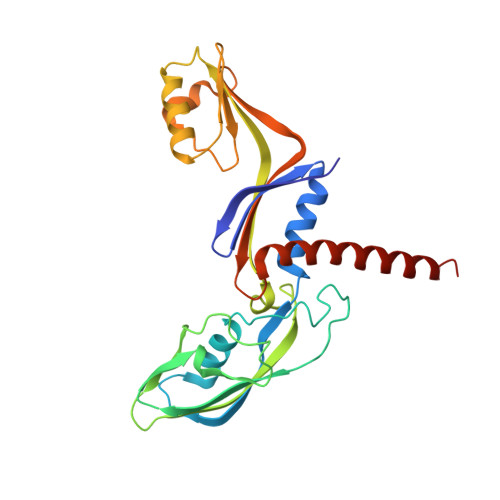
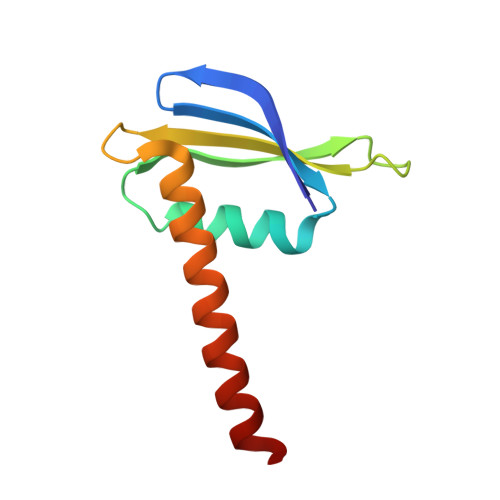
The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration
Jun, S.H., Hirata, A., Kanai, T., Santangelo, T.J., Imanaka, T., Murakami, K.S.(2014) Nat Commun 5: 5132-5132
- PubMed: 25311937
- DOI: https://doi.org/10.1038/ncomms6132
- Primary Citation of Related Structures:
4QIW, 4QJV - PubMed Abstract:
The archaeal transcription apparatus is closely related to the eukaryotic RNA polymerase II (Pol II) system. Archaeal RNA polymerase (RNAP) and Pol II evolved from a common ancestral structure and the euryarchaeal RNAP is the simplest member of the extant archaeal-eukaryotic RNAP family. Here we report the first crystal structure of euryarchaeal RNAP from Thermococcus kodakarensis (Tko). This structure reveals that the clamp domain is able to swing away from the main body of RNAP in the presence of the Rpo4/Rpo7 stalk by coordinated movements of these domains. More detailed structure-function analysis of yeast Pol II and Tko RNAP identifies structural additions to Pol II that correspond to the binding sites of Pol II-specific general transcription factors including TFIIF, TFIIH and Mediator. Such comparisons provide a framework for dissecting interactions between RNAP and these factors during formation of the pre-initiation complex.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The Center for RNA Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania 16802, USA.