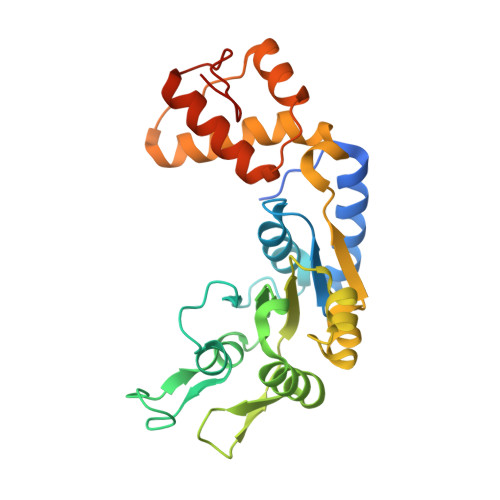
Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
Dey, S., Biswas, M., Sen, U., Dasgupta, J.(2015) J Biol Chem 290: 8734-8747
- PubMed: 25688103
- DOI: https://doi.org/10.1074/jbc.M114.611434
- Primary Citation of Related Structures:
4QHS, 4QHT - PubMed Abstract:
Bacterial enhancer-binding proteins (bEBPs) oligomerize through AAA(+) domains and use ATP hydrolysis-driven energy to isomerize the RNA polymerase-σ(54) complex during transcriptional initiation. Here, we describe the first structure of the central AAA(+) domain of the flagellar regulatory protein FlrC (FlrC(C)), a bEBP that controls flagellar synthesis in Vibrio cholerae. Our results showed that FlrC(C) forms heptamer both in nucleotide (Nt)-free and -bound states without ATP-dependent subunit remodeling. Unlike the bEBPs such as NtrC1 or PspF, a novel cis-mediated "all or none" ATP binding occurs in the heptameric FlrC(C), because constriction at the ATPase site, caused by loop L3 and helix α7, restricts the proximity of the trans-protomer required for Nt binding. A unique "closed to open" movement of Walker A, assisted by trans-acting "Glu switch" Glu-286, facilitates ATP binding and hydrolysis. Fluorescence quenching and ATPase assays on FlrC(C) and mutants revealed that although Arg-349 of sensor II, positioned by trans-acting Glu-286 and Tyr-290, acts as a key residue to bind and hydrolyze ATP, Arg-319 of α7 anchors ribose and controls the rate of ATP hydrolysis by retarding the expulsion of ADP. Heptameric state of FlrC(C) is restored in solution even with the transition state mimicking ADP·AlF3. Structural results and pulldown assays indicated that L3 renders an in-built geometry to L1 and L2 causing σ(54)-FlrC(C) interaction independent of Nt binding. Collectively, our results underscore a novel mechanism of ATP binding and σ(54) interaction that strives to understand the transcriptional mechanism of the bEBPs, which probably interact directly with the RNA polymerase-σ(54) complex without DNA looping.
Organizational Affiliation:
From the Department of Biotechnology, St. Xavier's College, 30 Park Street, Kolkata 700016 and.