Crystal structure of Leucurolysin-a complexed with an endogenous tripeptide (QSW).
Ferreira, R.N., Rates, B., Richardson, M., Guimaraes, B.G., Sanchez, E.O.F., Pimenta, A.M.C., Nagem, R.A.P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Snake venom metalloproteinase leucurolysin-A | 202 | Bothrops leucurus | Mutation(s): 0 EC: 3.4.24 | 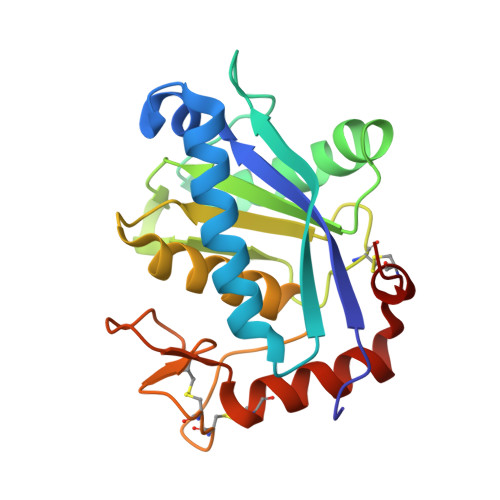 | |
UniProt | |||||
Find proteins for P84907 (Bothrops leucurus) Explore P84907 Go to UniProtKB: P84907 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P84907 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GLN-SER-TRP | B [auth D] | 3 | Bothrops leucurus | Mutation(s): 0 | 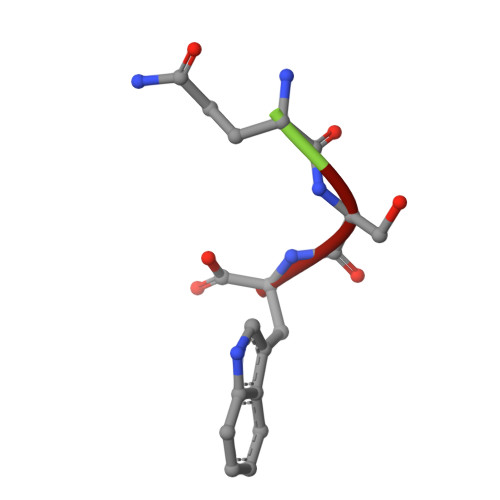 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | D [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | F [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | E [auth A] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| CA Query on CA | C [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 44.35 | α = 90 |
| b = 58.408 | β = 90 |
| c = 76.842 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASER | phasing |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |