Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2)
Bhukya, H., Bhujbalrao, R., Bitra, A., Anand, R.(2014) Nucleic Acids Res 42: 10122-10133
- PubMed: 25092919
- DOI: https://doi.org/10.1093/nar/gku587
- Primary Citation of Related Structures:
4PXI - PubMed Abstract:
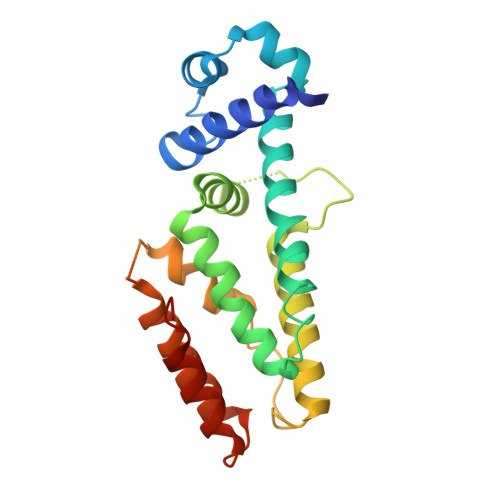
Antibiotic production and resistance pathways in Streptomyces are dictated by the interplay of transcriptional regulatory proteins that trigger downstream responses via binding to small diffusible molecules. To decipher the mode of DNA binding and the associated allosteric mechanism in the sub-class of transcription factors that are induced by γ-butyrolactones, we present the crystal structure of CprB in complex with the consensus DNA element to a resolution of 3.25 Å. Binding of the DNA results in the restructuring of the dimeric interface of CprB, inducing a pendulum-like motion of the helix-turn-helix motif that inserts into the major groove. The crystal structure revealed that, CprB is bound to DNA as a dimer of dimers with the mode of binding being analogous to the broad spectrum multidrug transporter protein QacR from the antibiotic resistant strain Staphylococcus aureus. It was demonstrated that the CprB displays a cooperative mode of DNA binding, following a clamp and click model. Experiments performed on a subset of DNA sequences from Streptomyces coelicolor A3(2) suggest that CprB is most likely a pleiotropic regulator. Apart from serving as an autoregulator, it is potentially a part of a network of proteins that modulates the γ-butyrolactone synthesis and antibiotic regulation pathways in S. coelicolor A3(2).
Organizational Affiliation:
Department of Chemistry, Indian Institute of Technology Bombay, Mumbai 400076, Maharashtra, India IITB-Monash Research Academy, Mumbai 400076, Maharashtra, India.