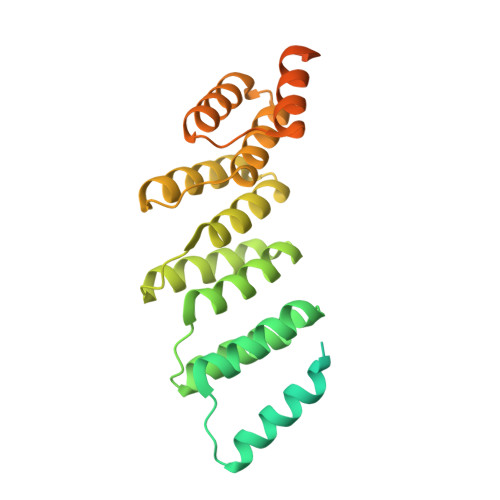
An artificial PPR scaffold for programmable RNA recognition.
Coquille, S., Filipovska, A., Chia, T., Rajappa, L., Lingford, J.P., Razif, M.F., Thore, S., Rackham, O.(2014) Nat Commun 5: 5729-5729
- PubMed: 25517350
- DOI: https://doi.org/10.1038/ncomms6729
- Primary Citation of Related Structures:
4PJQ, 4PJR, 4PJS, 4WN4, 4WSL - PubMed Abstract:
Pentatricopeptide repeat (PPR) proteins control diverse aspects of RNA metabolism in eukaryotic cells. Although recent computational and structural studies have provided insights into RNA recognition by PPR proteins, their highly insoluble nature and inconsistencies between predicted and observed modes of RNA binding have restricted our understanding of their biological functions and their use as tools. Here we use a consensus design strategy to create artificial PPR domains that are structurally robust and can be programmed for sequence-specific RNA binding. The atomic structures of these artificial PPR domains elucidate the structural basis for their stability and modelling of RNA-protein interactions provides mechanistic insights into the importance of RNA-binding residues and suggests modes of PPR-RNA association. The modular mode of RNA binding by PPR proteins holds great promise for the engineering of new tools to target RNA and to understand the mechanisms of gene regulation by natural PPR proteins.
Organizational Affiliation:
Department of Molecular Biology, University of Geneva, Science III, 30, Quai Ernest-Ansermet, Geneva 4 1211, Switzerland.