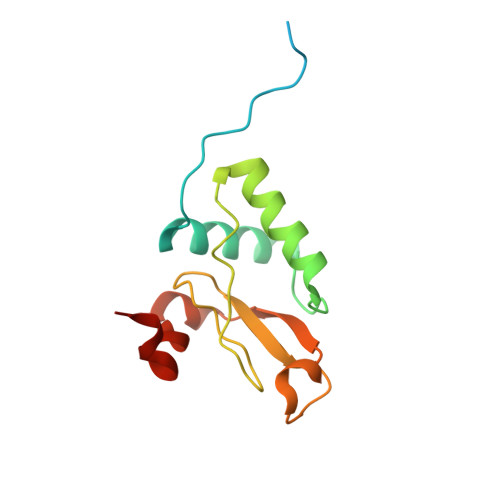
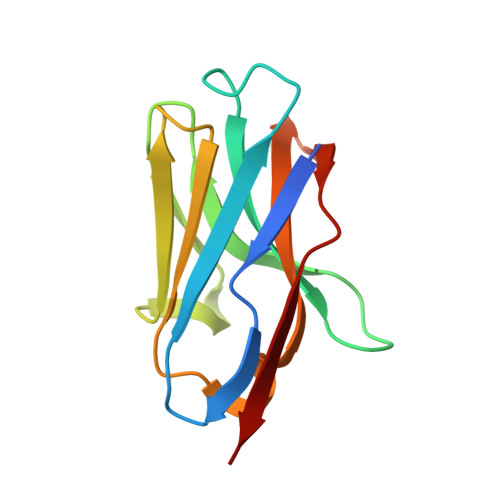
Structural basis for the inhibition of HIV-1 Nef by a high-affinity binding single-domain antibody.
Lulf, S., Matz, J., Rouyez, M.C., Jarviluoma, A., Saksela, K., Benichou, S., Geyer, M.(2014) Retrovirology 11: 24-24
- PubMed: 24620746
- DOI: https://doi.org/10.1186/1742-4690-11-24
- Primary Citation of Related Structures:
4ORZ - PubMed Abstract:
The HIV-1 Nef protein is essential for AIDS pathogenesis by its interaction with host cell surface receptors and signaling factors. Despite its critical role as a virulence factor Nef is not targeted by current antiviral strategies.
Organizational Affiliation:
Center of Advanced European Studies and Research, Group Physical Biochemistry, Bonn, Germany. matthias.geyer@caesar.de.