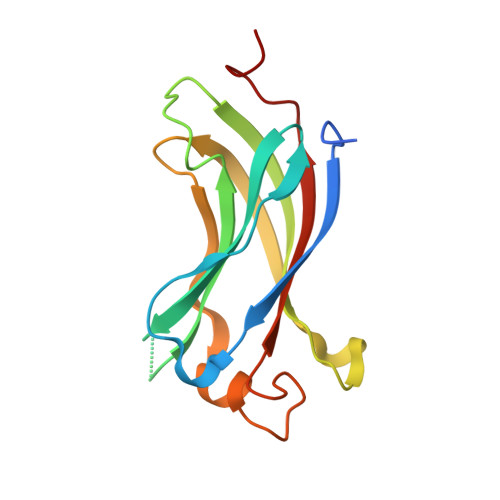
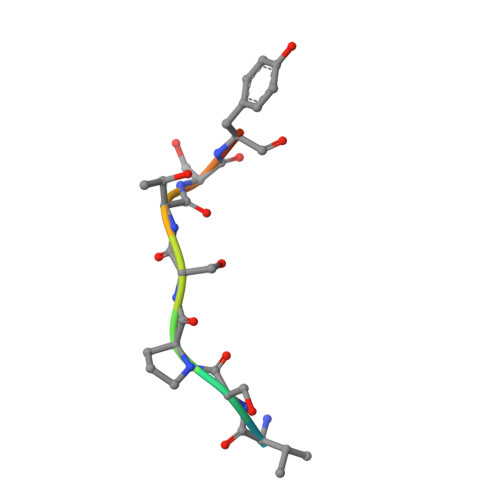
A role for USP7 in DNA replication.
Jagannathan, M., Nguyen, T., Gallo, D., Luthra, N., Brown, G.W., Saridakis, V., Frappier, L.(2014) Mol Cell Biol 34: 132-145
- PubMed: 24190967
- DOI: https://doi.org/10.1128/MCB.00639-13
- Primary Citation of Related Structures:
4KG9 - PubMed Abstract:
The minichromosome maintenance (MCM) complex, which plays multiple important roles in DNA replication, is loaded onto chromatin following mitosis, remains on chromatin until the completion of DNA synthesis, and then is unloaded by a poorly defined mechanism that involves the MCM binding protein (MCM-BP). Here we show that MCM-BP directly interacts with the ubiquitin-specific protease USP7, that this interaction occurs predominantly on chromatin, and that MCM-BP can tether USP7 to MCM proteins. Detailed biochemical and structure analyses of the USP7-MCM-BP interaction showed that the (155)PSTS(158) MCM-BP sequence mediates critical interactions with the TRAF domain binding pocket of USP7. Analysis of the effects of USP7 knockout on DNA replication revealed that lack of USP7 results in slowed progression through late S phase without globally affecting the fork rate or origin usage. Lack of USP7 also resulted in increased levels of MCM proteins on chromatin, and investigation of the cause of this increase revealed a defect in the dissociation of MCM proteins from chromatin in mid- to late S phase. This role of USP7 mirrors the previously described role for MCM-BP in MCM complex unloading and suggests that USP7 works with MCM-BP to unload MCM complexes from chromatin at the end of S phase.
Organizational Affiliation:
Department of Molecular Genetics, University of Toronto, Toronto, Canada.