Structural and Kinetic Characterization of OXA-45, a Class D beta-Lactamase with Extended Spectrum activity
Martin, J.D., Xiong, X.L., Catto, L.E., Toleman, M.A., Walsh, T.R., Clarke, A.R., Spencer, J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Oxacillinase | 240 | Pseudomonas aeruginosa | Mutation(s): 0 Gene Names: blaoxa45, oxa45 EC: 3.5.2.6 | 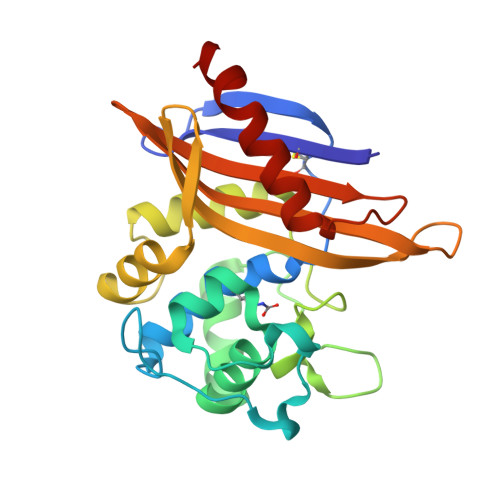 | |
UniProt | |||||
Find proteins for Q7WZC7 (Pseudomonas aeruginosa) Explore Q7WZC7 Go to UniProtKB: Q7WZC7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7WZC7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| KCX Query on KCX | A | L-PEPTIDE LINKING | C7 H14 N2 O4 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.169 | α = 90 |
| b = 47.488 | β = 90 |
| c = 108.903 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |