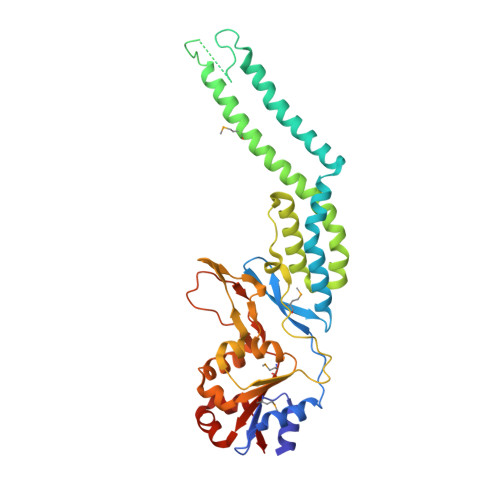
The N-terminal substrate-recognition domain of a LonC protease exhibits structural and functional similarity to cytosolic chaperones
Li, J.K., Liao, J.H., Li, H., Kuo, C.I., Huang, K.F., Yang, L.W., Wu, S.H., Chang, C.I.(2013) Acta Crystallogr D Biol Crystallogr 69: 1789-1797
- PubMed: 23999302
- DOI: https://doi.org/10.1107/S090744491301500X
- Primary Citation of Related Structures:
4FWV - PubMed Abstract:
The Lon protease is ubiquitous in nature. Its proteolytic activity is associated with diverse cellular functions ranging from maintaining proteostasis under normal and stress conditions to regulating cell metabolism. Although Lon was originally identified as an ATP-dependent protease with fused AAA+ (ATPases associated with diverse cellular activities) and protease domains, analyses have recently identified LonC as a class of Lon-like proteases with no intrinsic ATPase activity. In contrast to the canonical ATP-dependent Lon present in eukaryotic organelles and prokaryotes, LonC contains an AAA-like domain that lacks the conserved ATPase motifs. Moreover, the LonC AAA-like domain is inserted with a large domain predicted to be largely α-helical; intriguingly, this unique Lon-insertion domain (LID) was disordered in the recently determined full-length crystal structure of Meiothermus taiwanensis LonC (MtaLonC). Here, the crystal structure of the N-terminal AAA-like α/β subdomain of MtaLonC containing an intact LID, which forms a large α-helical hairpin protruding from the AAA-like domain, is reported. The structure of the LID is remarkably similar to the tentacle-like prong of the periplasmic chaperone Skp. It is shown that the LID of LonC is involved both in Skp-like chaperone activity and in recognition of unfolded protein substrates. The structure allows the construction of a complete model of LonC with six helical hairpin extensions defining a basket-like structure atop the AAA ring and encircling the entry portal to the barrel-like degradation chamber of Lon.
Organizational Affiliation:
Institute of Biochemical Sciences, College of Life Science, National Taiwan University, Taipei 10617, Taiwan.