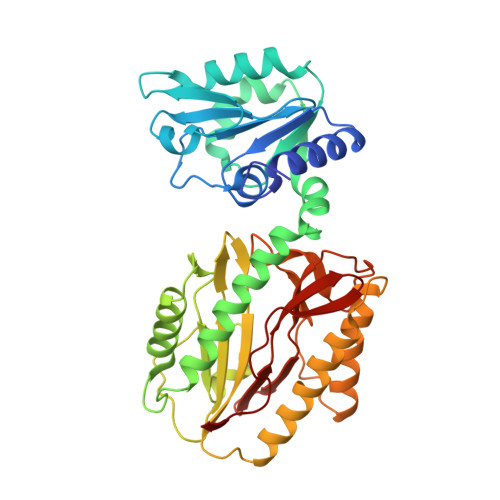
Influence of intermolecular contacts on the structure of recombinant prolidase from Thermococcus sibiricus.
Trofimov, A.A., Slutskaya, E.A., Polyakov, K.M., Dorovatovskii, P.V., Gumerov, V.M., Popov, V.O.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1275-1278
- PubMed: 23143231
- DOI: https://doi.org/10.1107/S174430911203761X
- Primary Citation of Related Structures:
4FKC - PubMed Abstract:
Prolidases are peptidases that are specific for dipeptides with proline as the second residue. The structure of recombinant prolidase from the hyperthermophilic archaeon Thermococcus sibiricus (Tsprol) was determined at 2.6 Å resolution. The homodimer of Tsprol is characterized by a complete lack of interactions between the N- and C-terminal domains of the two subunits and hence can be considered to be the most open structure when compared with previously structurally studied prolidases. This structure exists owing to intermolecular coordination bonds between cadmium ions derived from the crystallization solution and histidine residues of a His tag and aspartate and glutamate residues, which link the dimers to each other. This linking leads to the formation of a crystal with a loose packing of protein molecules and low resistance to mechanical influence and temperature increase.
Organizational Affiliation:
Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, ul. Vavilova 32, Moscow 119991, Russian Federation. burayatina@gmail.com