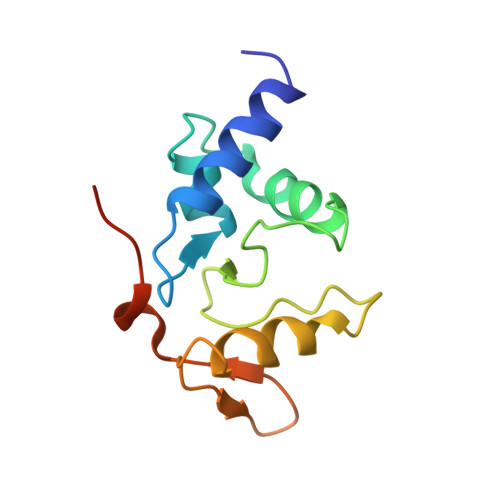
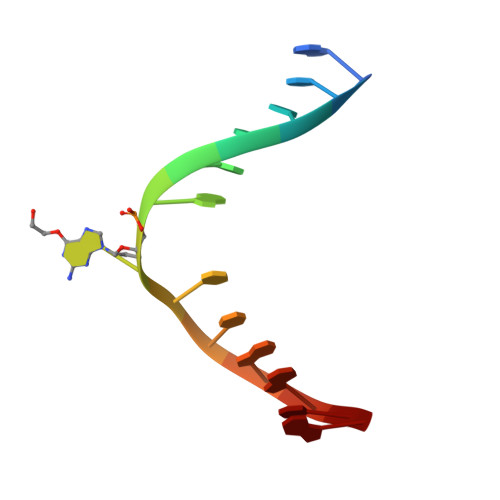
Atl1 Regulates Choice between Global Genome and Transcription-Coupled Repair of O(6)-Alkylguanines.
Latypov, V.F., Tubbs, J.L., Watson, A.J., Marriott, A.S., McGown, G., Thorncroft, M., Wilkinson, O.J., Senthong, P., Butt, A., Arvai, A.S., Millington, C.L., Povey, A.C., Williams, D.M., Santibanez-Koref, M.F., Tainer, J.A., Margison, G.P.(2012) Mol Cell 47: 50-60
- PubMed: 22658721
- DOI: https://doi.org/10.1016/j.molcel.2012.04.028
- Primary Citation of Related Structures:
4ENJ, 4ENK, 4ENM, 4ENN - PubMed Abstract:
Nucleotide excision repair (NER) has long been known to remove DNA lesions induced by chemical carcinogens, and the molecular mechanism has been partially elucidated. Here we demonstrate that in Schizosaccharomyces pombe a DNA recognition protein, alkyltransferase-like 1 (Atl1), can play a pivotal role in selecting a specific NER pathway, depending on the nature of the DNA modification. The relative ease of dissociation of Atl1 from DNA containing small O(6)-alkylguanines allows accurate completion of global genome repair (GGR), whereas strong Atl1 binding to bulky O(6)-alkylguanines blocks GGR, stalls the transcription machinery, and diverts the damage to transcription-coupled repair. Our findings redraw the initial stages of the NER process in those organisms that express an alkyltransferase-like gene and raise the question of whether or not O(6)-alkylguanine lesions that are poor substrates for the alkyltransferase proteins in higher eukaryotes might, by analogy, signal such lesions for repair by NER.
Organizational Affiliation:
Cancer Research UK Carcinogenesis Group, Paterson Institute for Cancer Research, University of Manchester, Manchester M20 4BX, UK.