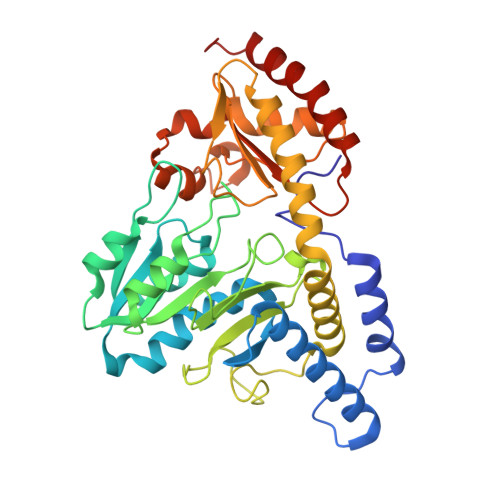
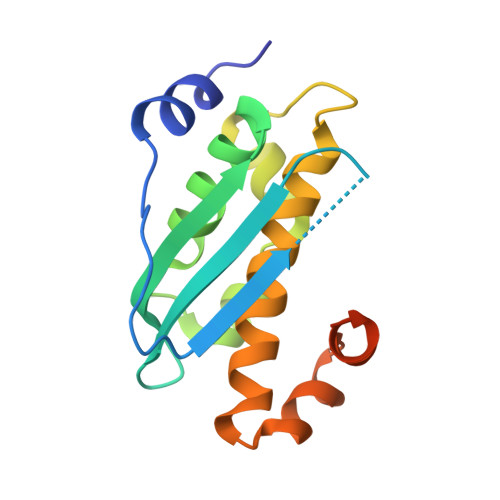
(IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer.
Marinoni, E.N., de Oliveira, J.S., Nicolet, Y., Raulfs, E.C., Amara, P., Dean, D.R., Fontecilla-Camps, J.C.(2012) Angew Chem Int Ed Engl 51: 5439-5442
- PubMed: 22511353
- DOI: https://doi.org/10.1002/anie.201201708
- Primary Citation of Related Structures:
4EB5, 4EB7
Organizational Affiliation:
Metalloproteins Unit, Institut de Biologie Structurale J. P. Ebel, Commissariat à l'Énergie Atomique, Centre National de la Recherche Scientifique, Université Joseph Fourier, Grenoble, France.