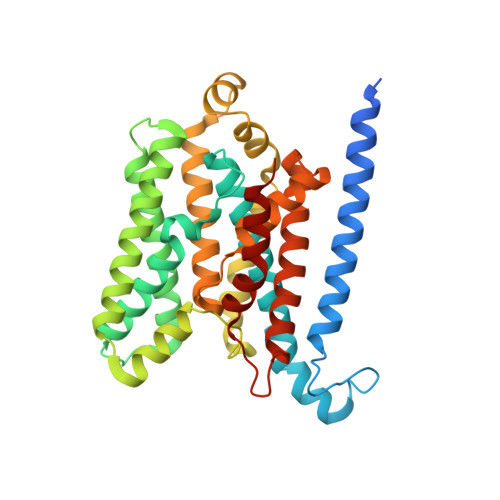
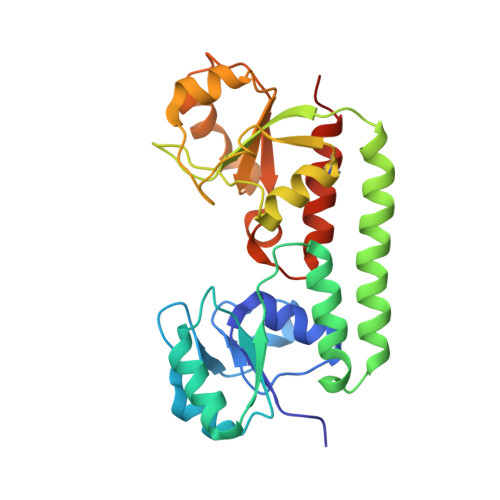
Asymmetric states of vitamin B12 transporter BtuCD are not discriminated by its cognate substrate binding protein BtuF.
Korkhov, V.M., Mireku, S.A., Hvorup, R.N., Locher, K.P.(2012) FEBS Lett 586: 972-976
- PubMed: 22569249
- DOI: https://doi.org/10.1016/j.febslet.2012.02.042
- Primary Citation of Related Structures:
4DBL - PubMed Abstract:
BtuCD is an ABC transporter catalyzing the uptake of vitamin B₁₂ across the Escherichia coli inner membrane. A previously reported X-ray structure of BtuCD in complex with the periplasmic vitamin B₁₂-binding protein BtuF revealed asymmetry of the transmembrane BtuC subunits. The functional relevance of this asymmetry has remained uncertain. Here we report the X-ray structure of a catalytically impaired BtuCD mutant in complex with BtuF, where the BtuC subunits adopt a distinct asymmetric conformation. The structure suggests that BtuF does not discriminate between, or impose, asymmetric conformations of BtuCD. It also explains the conformational disorder observed in BtuCDF crystals.
Organizational Affiliation:
Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich, Switzerland.