The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
Lee, E.F., Dewson, G., Evangelista, M., Pettikiriarachchi, A., Zhu, H., Colman, P.M., Fairlie, W.D.(2014) J Biol Chem 289: 36001
- PubMed: 25371206
- DOI: https://doi.org/10.1074/jbc.M114.610758
- Primary Citation of Related Structures:
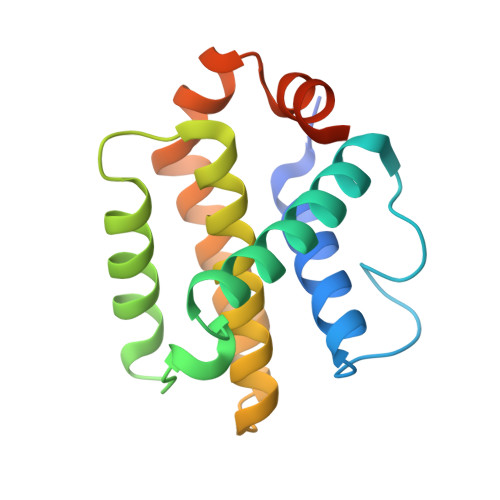
4CIM, 4CIN - PubMed Abstract:
Bcl-2 homology 3 (BH3) domains are short sequence motifs that mediate nearly all protein-protein interactions between B cell lymphoma 2 (Bcl-2) family proteins in the intrinsic apoptotic cell death pathway. These sequences are found on both pro-survival and pro-apoptotic members, although their primary function is believed to be associated with induction of cell death. Here, we identify critical features of the BH3 domains of pro-survival proteins that distinguish them functionally from their pro-apoptotic counterparts. Biochemical and x-ray crystallographic studies demonstrate that these differences reduce the capacity of most pro-survival proteins to form high affinity "BH3-in-groove" complexes that are critical for cell death induction. Switching these residues for the corresponding residues in Bcl-2 homologous antagonist/killer (Bak) increases the binding affinity of isolated BH3 domains for pro-survival proteins; however, their exchange in the context of the parental protein causes rapid proteasomal degradation due to protein destabilization. This is supported by further x-ray crystallographic studies that capture elements of this destabilization in one pro-survival protein, Bcl-w. In pro-apoptotic Bak, we demonstrate that the corresponding distinguishing residues are important for its cell-killing capacity and antagonism by pro-survival proteins.
Organizational Affiliation:
From the Walter and Eliza Hall Institute of Medical Research, 1G Royal Pde, Parkville, Victoria 3052, Australia and the Department of Medical Biology, University of Melbourne, Parkville, Victoria 3010, Australia.