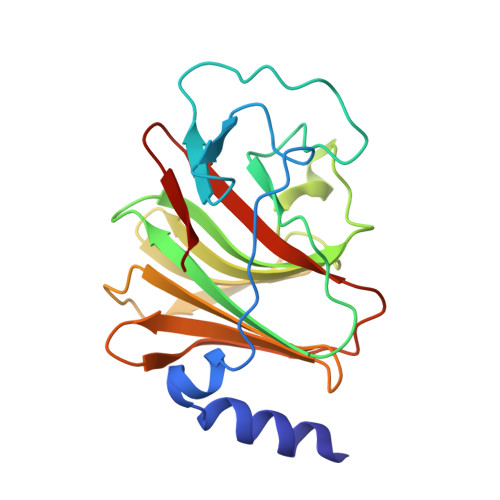
Crystal Structure of the Trim25 B30.2 (Pryspry) Domain: A Key Component of Antiviral Signaling.
D'Cruz, A.A., Kershaw, N.J., Chiang, J.J., Wang, M.K., Nicola, N.A., Babon, J.J., Gack, M.U., Nicholson, S.E.(2013) Biochem J 456: 231
- PubMed: 24015671
- DOI: https://doi.org/10.1042/BJ20121425
- Primary Citation of Related Structures:
4B8E - PubMed Abstract:
TRIM (tripartite motif) proteins primarily function as ubiquitin E3 ligases that regulate the innate immune response to infection. TRIM25 [also known as Efp (oestrogen-responsive finger protein)] has been implicated in the regulation of oestrogen receptor α signalling and in the regulation of innate immune signalling via RIG-I (retinoic acid-inducible gene-I). RIG-I senses cytosolic viral RNA and is subsequently ubiquitinated by TRIM25 at its N-terminal CARDs (caspase recruitment domains), leading to type I interferon production. The interaction with RIG-I is dependent on the TRIM25 B30.2 domain, a protein-interaction domain composed of the PRY and SPRY tandem sequence motifs. In the present study we describe the 1.8 Å crystal structure of the TRIM25 B30.2 domain, which exhibits a typical B30.2/SPRY domain fold comprising two N-terminal α-helices, thirteen β-strands arranged into two β-sheets and loop regions of varying lengths. A comparison with other B30.2/SPRY structures and an analysis of the loop regions identified a putative binding pocket, which is likely to be involved in binding target proteins. This was supported by mutagenesis and functional analyses, which identified two key residues (Asp(488) and Trp(621)) in the TRIM25 B30.2 domain as being critical for binding to the RIG-I CARDs.
Organizational Affiliation:
‡Department of Microbiology and Immunobiology, Harvard Medical School, Boston, MA, U.S.A.