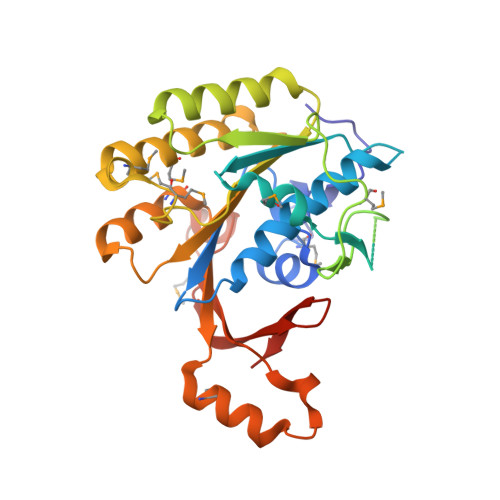
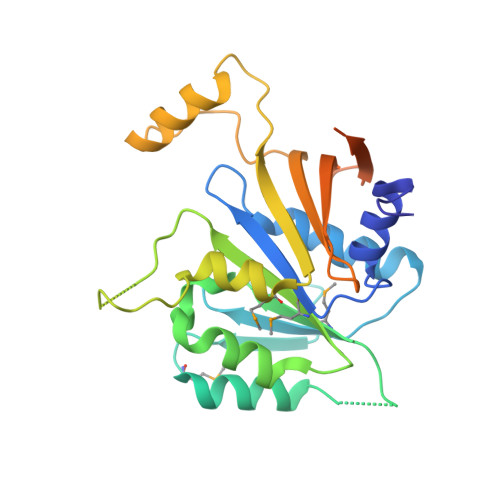
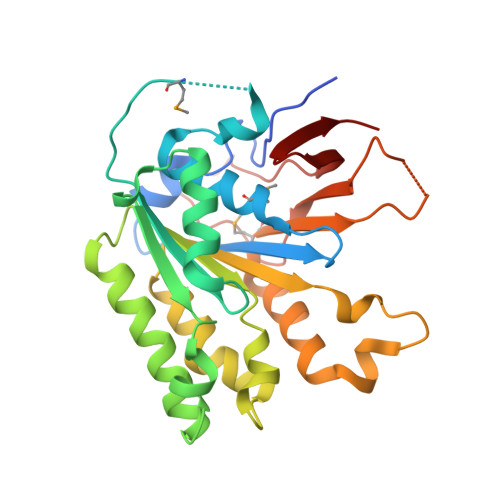
The Elongator Subcomplex Elp456 is a Hexameric Reca-Like ATPase.
Glatt, S., Letoquart, J., Faux, C., Taylor, N.M.I., Seraphin, B., Muller, C.W.(2012) Nat Struct Mol Biol 19: 314
- PubMed: 22343726
- DOI: https://doi.org/10.1038/nsmb.2234
- Primary Citation of Related Structures:
4A8J - PubMed Abstract:
Elongator was initially described as an RNA polymerase II-associated factor but has since been associated with a broad range of cellular activities. It has also attracted clinical attention because of its role in certain neurodegenerative diseases. Here we describe the crystal structure of the Saccharomyces cerevisiae subcomplex of Elongator proteins 4, 5 and 6 (Elp456). The subunits each show almost identical RecA folds that form a heterohexameric ring-like structure resembling hexameric RecA-like ATPases. This structural finding is supported by different complementary in vitro and in vivo approaches, including the specific binding of the hexameric Elp456 subcomplex to tRNAs in a manner regulated by ATP. Our results support a role of Elongator in tRNA modification, explain the importance of each of the Elp4, Elp5 and Elp6 subunits for complex integrity and suggest a model for the overall architecture of the holo-Elongator complex.
Organizational Affiliation:
European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Heidelberg, Germany.