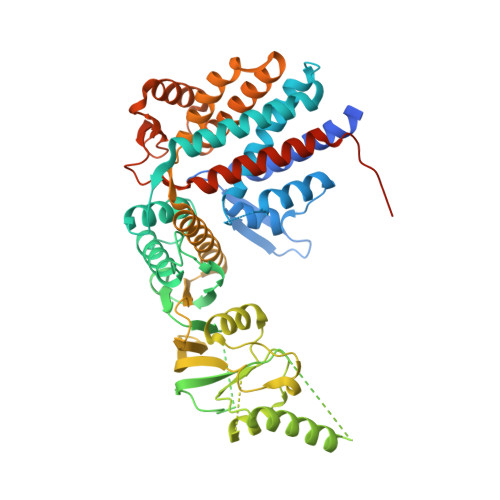
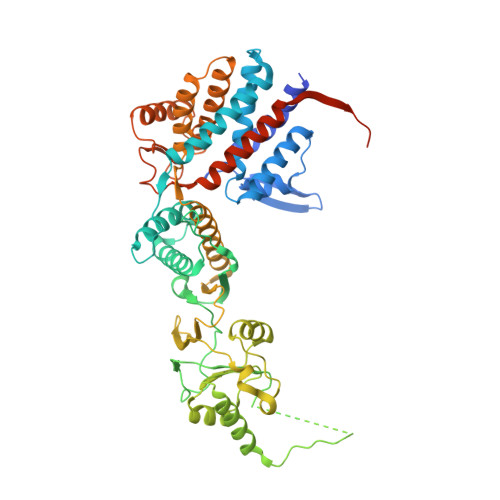
Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Chaston, J.J., Smits, C., Aragao, D., Wong, A.S., Ahsan, B., Sandin, S., Molugu, S.K., Molugu, S.K., Bernal, R.A., Stock, D., Stewart, A.G.(2016) Structure 24: 364-374
- PubMed: 26853941
- DOI: https://doi.org/10.1016/j.str.2015.12.016
- Primary Citation of Related Structures:
4XCD, 4XCG, 4XCI - PubMed Abstract:
Chaperonins are essential biological complexes assisting protein folding in all kingdoms of life. Whereas homooligomeric bacterial GroEL binds hydrophobic substrates non-specifically, the heterooligomeric eukaryotic CCT binds specifically to distinct classes of substrates. Sulfolobales, which survive in a wide range of temperatures, have evolved three different chaperonin subunits (α, β, γ) that form three distinct complexes tailored for different substrate classes at cold, normal, and elevated temperatures. The larger octadecameric β complexes cater for substrates under heat stress, whereas smaller hexadecameric αβ complexes prevail under normal conditions. The cold-shock complex contains all three subunits, consistent with greater substrate specificity. Structural analysis using crystallography and electron microscopy reveals the geometry of these complexes and shows a novel arrangement of the α and β subunits in the hexadecamer enabling incorporation of the γ subunit.
Organizational Affiliation:
Molecular, Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Darlinghurst, NSW 2010, Australia; Faculty of Medicine, The University of New South Wales, Sydney, NSW 2052, Australia.