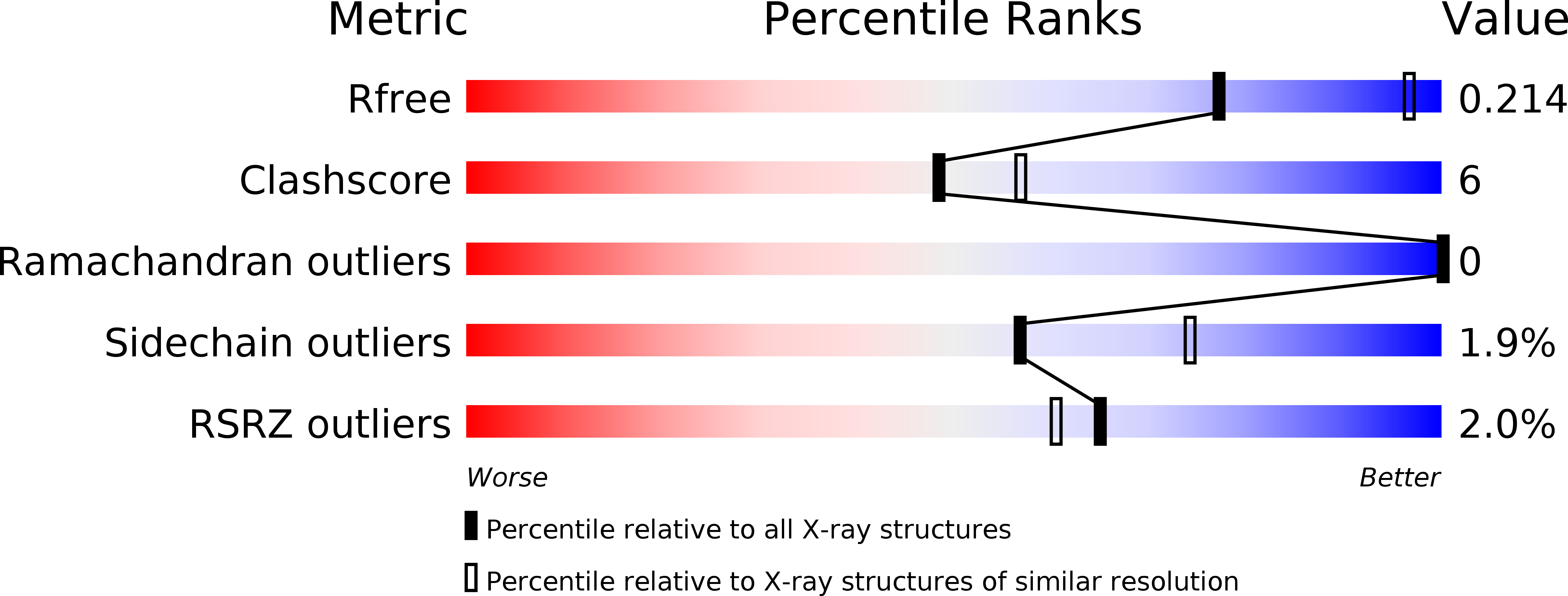
A Switch between One- and Two-electron Chemistry of the Human Flavoprotein Iodotyrosine Deiodinase Is Controlled by Substrate.
Hu, J., Chuenchor, W., Rokita, S.E.(2015) J Biol Chem 290: 590-600
- PubMed: 25395621
- DOI: https://doi.org/10.1074/jbc.M114.605964
- Primary Citation of Related Structures:
4TTB, 4TTC - PubMed Abstract:
Reductive dehalogenation is not typical of aerobic organisms but plays a significant role in iodide homeostasis and thyroid activity. The flavoprotein iodotyrosine deiodinase (IYD) is responsible for iodide salvage by reductive deiodination of the iodotyrosine derivatives formed as byproducts of thyroid hormone biosynthesis. Heterologous expression of the human enzyme lacking its N-terminal membrane anchor has allowed for physical and biochemical studies to identify the role of substrate in controlling the active site geometry and flavin chemistry. Crystal structures of human IYD and its complex with 3-iodo-l-tyrosine illustrate the ability of the substrate to provide multiple interactions with the isoalloxazine system of FMN that are usually provided by protein side chains. Ligand binding acts to template the active site geometry and significantly stabilize the one-electron-reduced semiquinone form of FMN. The neutral form of this semiquinone is observed during reductive titration of IYD in the presence of the substrate analog 3-fluoro-l-tyrosine. In the absence of an active site ligand, only the oxidized and two-electron-reduced forms of FMN are detected. The pH dependence of IYD binding and turnover also supports the importance of direct coordination between substrate and FMN for productive catalysis.
Organizational Affiliation:
From the Department of Chemistry and Biochemistry, University of Maryland, College Park, Maryland 20742 and.