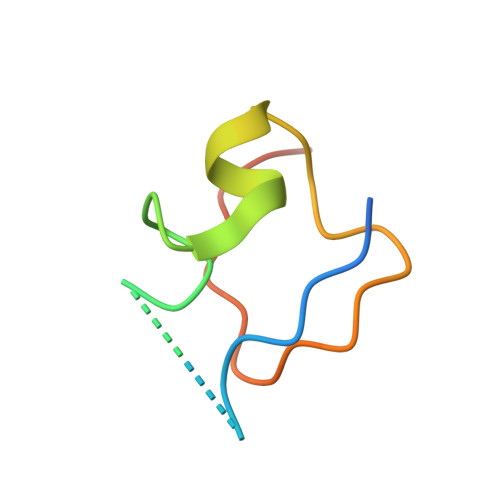
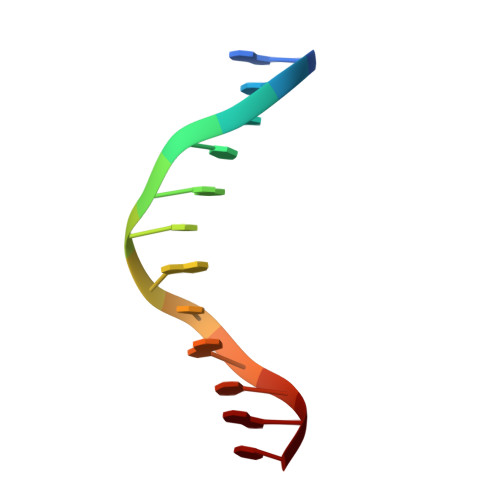
Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Zheng, S., Villa, R., Wang, J., Feng, Y., Wang, J., Becker, P.B., Ye, K.(2014) Genes Dev 28: 2652-2662
- PubMed: 25452275
- DOI: https://doi.org/10.1101/gad.250936.114
- Primary Citation of Related Structures:
4RKG, 4RKH - PubMed Abstract:
The male-specific lethal dosage compensation complex (MSL-DCC) selectively assembles on the X chromosome in Drosophila males and activates gene transcription by twofold through histone acetylation. An MSL recognition element (MRE) sequence motif nucleates the initial MSL association, but how it is recognized remains unknown. Here, we identified the CXC domain of MSL2 specifically recognizing the MRE motif and determined its crystal structure bound to specific and nonspecific DNAs. The CXC domain primarily contacts one strand of DNA duplex and employs a single arginine to directly read out dinucleotide sequences from the minor groove. The arginine is flexible when bound to nonspecific sequences. The core region of the MRE motif harbors two binding sites on opposite strands that can cooperatively recruit a CXC dimer. Specific DNA-binding mutants of MSL2 are impaired in MRE binding and X chromosome localization in vivo. Our results reveal multiple dynamic DNA-binding modes of the CXC domain that target the MSL-DCC to X chromosomes.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, College of Life Sciences, Beijing Normal University, Beijing 100875, China; National Institute of Biological Sciences, Beijing 102206, China;