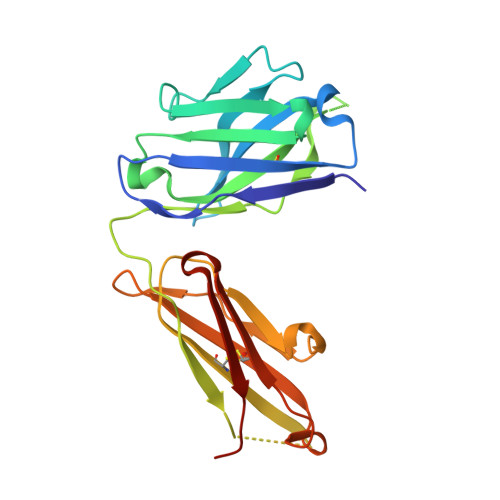
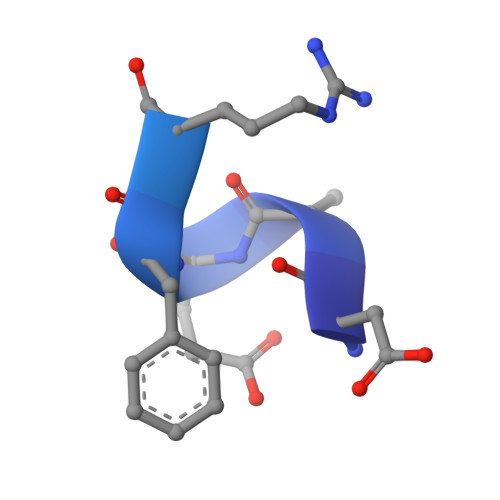
Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Feinberg, H., Saldanha, J.W., Diep, L., Goel, A., Widom, A., Veldman, G.M., Weis, W.I., Schenk, D., Basi, G.S.(2014) Alzheimers Res Ther 6: 31-31
- PubMed: 25024748
- DOI: https://doi.org/10.1186/alzrt261
- Primary Citation of Related Structures:
4ONF, 4ONG - PubMed Abstract:
Immunotherapy targeting amyloid-β peptide is under active clinical investigation for treatment of Alzheimer's disease (AD). Among the hypotheses being investigated for impact on clinical outcome are the preferred epitope or conformation of amyloid-β to target for treatment, and the mechanism of action underlying immunotherapy. Bapineuzumab (humanized 3D6), a neo-epitope specific antibody recognizing amyloid-β1-5 with strong preference for an exposed Asp residue at the N-terminus of the peptide, has undergone advanced clinical testing for treatment of AD.
Organizational Affiliation:
Departments of Structural Biology and of Molecular & Cellular Physiology, 299 Campus Drive, Stanford University School of Medicine, Stanford, CA 94305, USA.