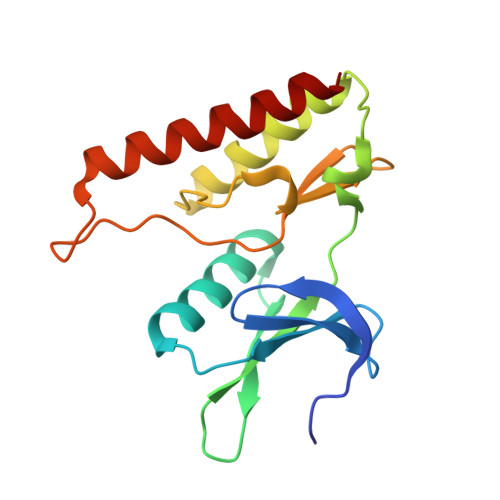
Type III Effector NleH2 from Escherichia coli O157:H7 str. Sakai Features an Atypical Protein Kinase Domain.
Halavaty, A.S., Anderson, S.M., Wawrzak, Z., Kudritska, M., Skarina, T., Anderson, W.F., Savchenko, A.(2014) Biochemistry 53: 2433-2435
- PubMed: 24712300
- DOI: https://doi.org/10.1021/bi500016j
- Primary Citation of Related Structures:
4O96 - PubMed Abstract:
The crystal structure of a C-terminal domain of enterohemorrhagic Escherichia coli type III effector NleH2 has been determined to 2.6 Å resolution. The structure resembles those of protein kinases featuring the catalytic, activation, and glycine-rich loop motifs and ATP-binding site. The position of helix αC and the lack of a conserved arginine within an equivalent HRD motif suggested that the NleH2 kinase domain's active conformation might not require phosphorylation. The activation segment markedly contributed to the dimerization interface of NleH2, which can also accommodate the NleH1-NleH2 heterodimer. The C-terminal PDZ-binding motif of NleH2 provided bases for interaction with host proteins.
Organizational Affiliation:
Center for Structural Genomics of Infectious Diseases (CSGID), Molecular Pharmacology and Biological Chemistry, Northwestern University , Chicago, Illinois 60611, United States.