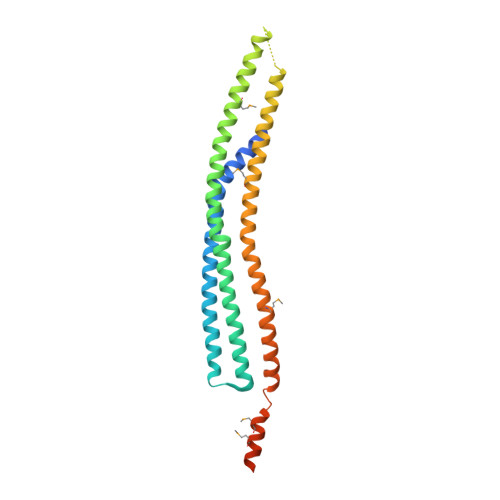
The inverse BAR domain protein IBARa drives membrane remodeling to control osmoregulation, phagocytosis and cytokinesis.
Linkner, J., Witte, G., Zhao, H., Junemann, A., Nordholz, B., Runge-Wollmann, P., Lappalainen, P., Faix, J.(2014) J Cell Sci 127: 1279-1292
- PubMed: 24463811
- DOI: https://doi.org/10.1242/jcs.140756
- Primary Citation of Related Structures:
4NQI - PubMed Abstract:
Here, we analyzed the single inverse Bin/Amphiphysin/Rvs (I-BAR) family member IBARa from Dictyostelium discoideum. The X-ray structure of the N-terminal I-BAR domain solved at 2.2 Å resolution revealed an all-α-helical structure that self-associates into a 165-Å zeppelin-shaped antiparallel dimer. The structural data are consistent with its shape in solution obtained by small-angle X-ray scattering. Cosedimentation, fluorescence anisotropy, and fluorescence and electron microscopy revealed that the I-BAR domain bound preferentially to phosphoinositide-containing vesicles and drove the formation of negatively curved tubules. Immunofluorescence labeling further showed accumulation of endogenous IBARa at the tips of filopodia, the rim of constricting phagocytic cups, in foci connecting dividing cells during the final stage of cytokinesis and most prominently at the osmoregulatory contractile vacuole (CV). Consistently, IBARa-null mutants displayed defects in CV formation and discharge, growth, phagocytosis and mitotic cell division, whereas filopodia formation was not compromised. Of note, IBARa-null mutants were also strongly impaired in cell spreading. Taken together, these data suggest that IBARa constitutes an important regulator of numerous cellular processes intimately linked with the dynamic rearrangement of cellular membranes.
Organizational Affiliation:
Institute for Biophysical Chemistry, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany.