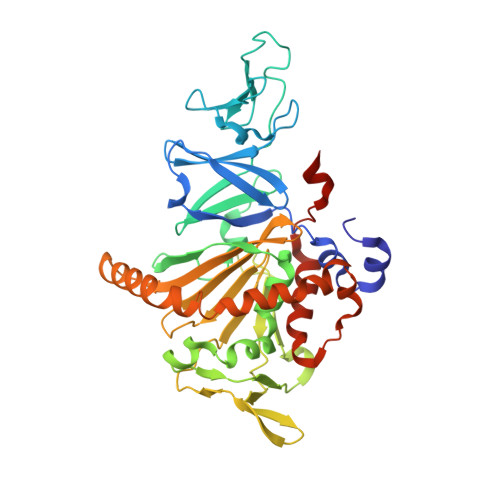
Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Inoue, K., Usami, Y., Ashikawa, Y., Noguchi, H., Umeda, T., Yamagami-Ashikawa, A., Horisaki, T., Uchimura, H., Terada, T., Nakamura, S., Shimizu, K., Habe, H., Yamane, H., Fujimoto, Z., Nojiri, H.(2014) Appl Environ Microbiol 80: 2821-2832
- PubMed: 24584240
- DOI: https://doi.org/10.1128/AEM.04000-13
- Primary Citation of Related Structures:
4NB8, 4NB9, 4NBA, 4NBB, 4NBC, 4NBD, 4NBE, 4NBF, 4NBG, 4NBH - PubMed Abstract:
Carbazole 1,9a-dioxygenase (CARDO), a Rieske nonheme iron oxygenase (RO), is a three-component system composed of a terminal oxygenase (Oxy), ferredoxin, and a ferredoxin reductase. Oxy has angular dioxygenation activity against carbazole. Previously, site-directed mutagenesis of the Oxy-encoding gene from Janthinobacterium sp. strain J3 generated the I262V, F275W, Q282N, and Q282Y Oxy derivatives, which showed oxygenation capabilities different from those of the wild-type enzyme. To understand the structural features resulting in the different oxidation reactions, we determined the crystal structures of the derivatives, both free and complexed with substrates. The I262V, F275W, and Q282Y derivatives catalyze the lateral dioxygenation of carbazole with higher yields than the wild type. A previous study determined the crystal structure of Oxy complexed with carbazole and revealed that the carbonyl oxygen of Gly178 hydrogen bonds with the imino nitrogen of carbazole. In these derivatives, the carbazole was rotated approximately 15, 25, and 25°, respectively, compared to the wild type, creating space for a water molecule, which hydrogen bonds with the carbonyl oxygen of Gly178 and the imino nitrogen of carbazole. In the crystal structure of the F275W derivative complexed with fluorene, C-9 of fluorene, which corresponds to the imino nitrogen of carbazole, was oriented close to the mutated residue Trp275, which is on the opposite side of the binding pocket from the carbonyl oxygen of Gly178. Our structural analyses demonstrate that the fine-tuning of hydrophobic residues on the surface of the substrate-binding pocket in ROs causes a slight shift in the substrate-binding position that, in turn, favors specific oxygenation reactions toward various substrates.
Organizational Affiliation:
Biotechnology Research Center, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.