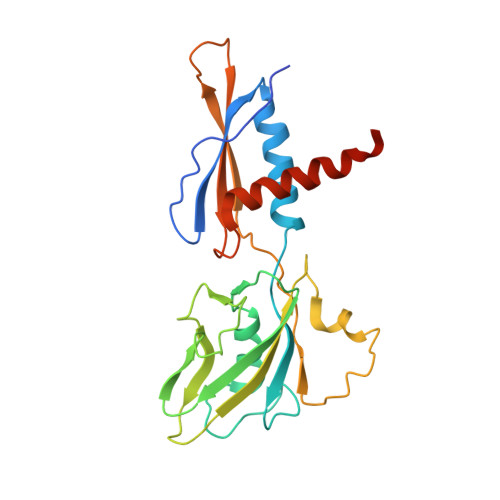
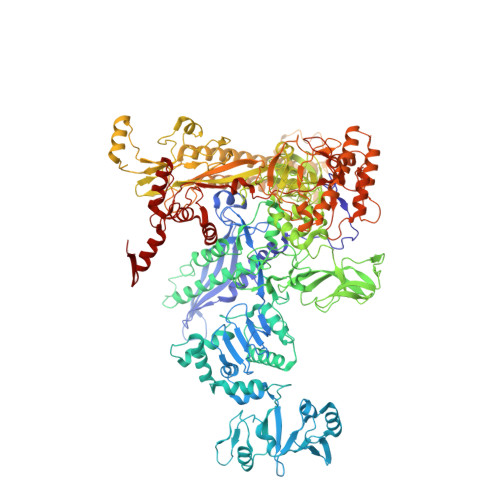
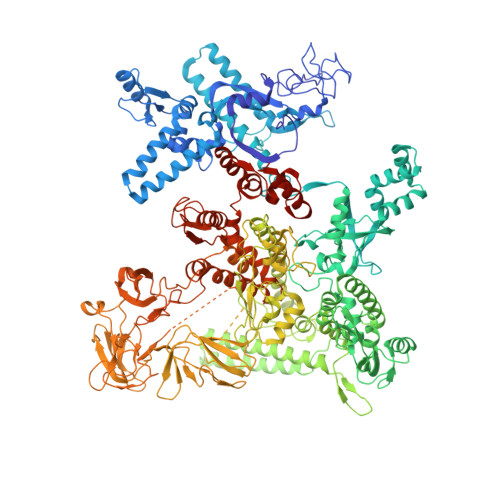
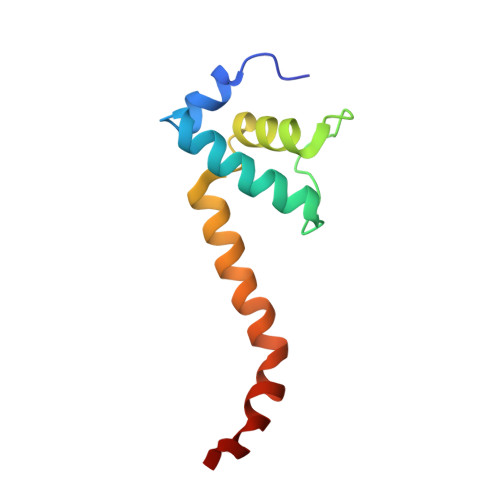
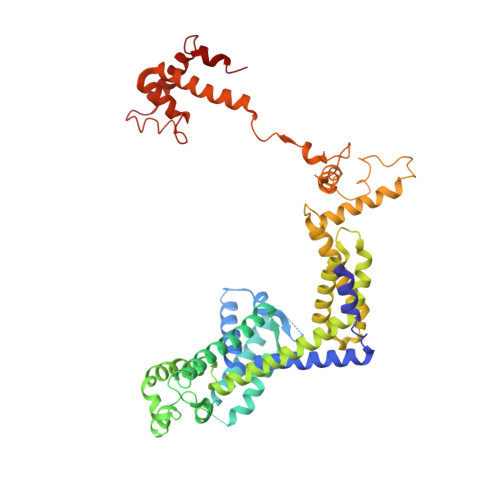
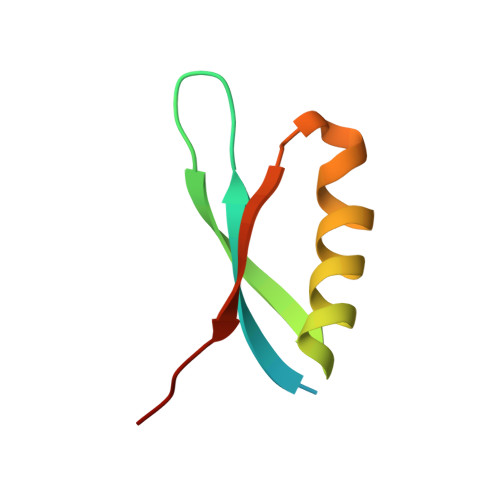
Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Bae, B., Davis, E., Brown, D., Campbell, E.A., Wigneshweraraj, S., Darst, S.A.(2013) Proc Natl Acad Sci U S A 110: 19772-19777
- PubMed: 24218560
- DOI: https://doi.org/10.1073/pnas.1314576110
- Primary Citation of Related Structures:
4LJZ, 4LK0, 4LK1, 4LLG - PubMed Abstract:
Bacteriophage T7 encodes an essential inhibitor of the Escherichia coli host RNA polymerase (RNAP), the product of gene 2 (Gp2). We determined a series of X-ray crystal structures of E. coli RNAP holoenzyme with or without Gp2. The results define the structure and location of the RNAP σ(70) subunit domain 1.1(σ(1.1)(70)) inside the RNAP active site channel, where it must be displaced by the DNA upon formation of the open promoter complex. The structures and associated data, combined with previous results, allow for a complete delineation of the mechanism for Gp2 inhibition of E. coli RNAP. In the primary inhibition mechanism, Gp2 forms a protein-protein interaction with σ(1.1)(70), preventing the normal egress of σ(1.1)(70) from the RNAP active site channel. Gp2 thus misappropriates a domain of the RNAP holoenzyme, σ(1.1)(70), to inhibit the function of the enzyme.
Organizational Affiliation:
Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY 10065.