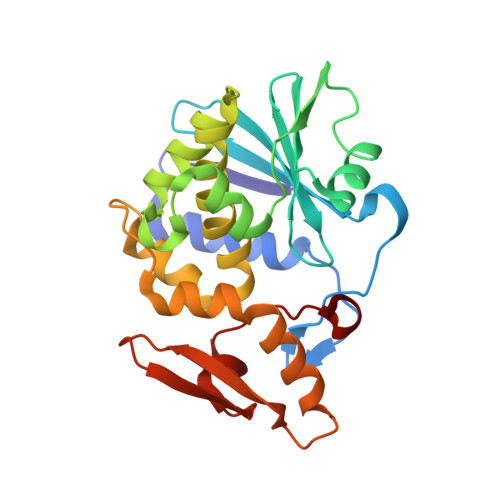
Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Polyethylene glycol at 1.90 Angstrom resolution
Pandey, S., Tyagi, T.K., Singh, A., Bhushan, A., Kushwaha, G.S., Sinha, M., Kaur, P., Sharma, S., Singh, T.P.To be published.