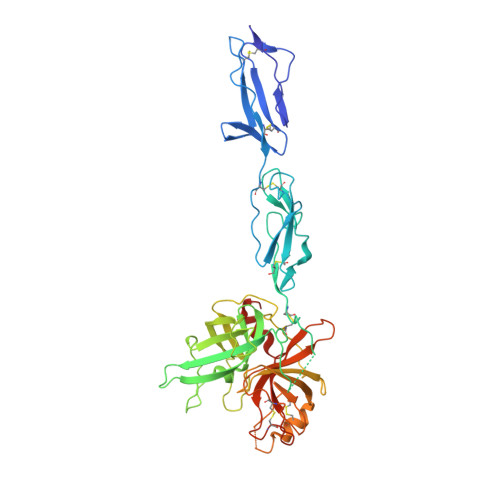
The X-ray Crystal Structure of Mannose-binding Lectin-associated Serine Proteinase-3 Reveals the Structural Basis for Enzyme Inactivity Associated with the Carnevale, Mingarelli, Malpuech, and Michels (3MC) Syndrome.
Yongqing, T., Wilmann, P.G., Reeve, S.B., Coetzer, T.H., Smith, A.I., Whisstock, J.C., Pike, R.N., Wijeyewickrema, L.C.(2013) J Biol Chem 288: 22399-22407
- PubMed: 23792966
- DOI: https://doi.org/10.1074/jbc.M113.483875
- Primary Citation of Related Structures:
4KKD - PubMed Abstract:
The mannose-binding lectin associated-protease-3 (MASP-3) is a member of the lectin pathway of the complement system, a key component of human innate and active immunity. Mutations in MASP-3 have recently been found to be associated with Carnevale, Mingarelli, Malpuech, and Michels (3MC) syndrome, a severe developmental disorder manifested by cleft palate, intellectual disability, and skeletal abnormalities. However, the molecular basis for MASP-3 function remains to be understood. Here we characterize the substrate specificity of MASP-3 by screening against a combinatorial peptide substrate library. Through this approach, we successfully identified a peptide substrate that was 20-fold more efficiently cleaved than any other identified to date. Furthermore, we demonstrated that mutant forms of the enzyme associated with 3MC syndrome were completely inactive against this substrate. To address the structural basis for this defect, we determined the 2.6-Å structure of the zymogen form of the G666E mutant of MASP-3. These data reveal that the mutation disrupts the active site and perturbs the position of the catalytic serine residue. Together, these insights into the function of MASP-3 reveal how a mutation in this enzyme causes it to be inactive and thus contribute to the 3MC syndrome.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology, Monash University, Clayton, Victoria 3800, Australia.