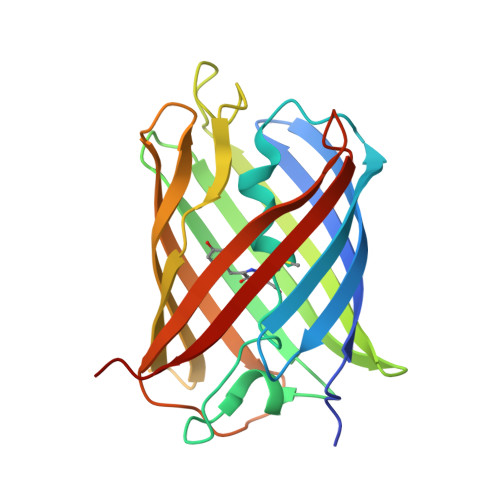
Split green fluorescent protein as a modular binding partner for protein crystallization.
Nguyen, H.B., Hung, L.W., Yeates, T.O., Terwilliger, T.C., Waldo, G.S.(2013) Acta Crystallogr D Biol Crystallogr 69: 2513-2523
- PubMed: 24311592
- DOI: https://doi.org/10.1107/S0907444913024608
- Primary Citation of Related Structures:
4KF4, 4KF5 - PubMed Abstract:
A modular strategy for protein crystallization using split green fluorescent protein (GFP) as a crystallization partner is demonstrated. Insertion of a hairpin containing GFP β-strands 10 and 11 into a surface loop of a target protein provides two chain crossings between the target and the reconstituted GFP compared with the single connection afforded by terminal GFP fusions. This strategy was tested by inserting this hairpin into a loop of another fluorescent protein, sfCherry. The crystal structure of the sfCherry-GFP(10-11) hairpin in complex with GFP(1-9) was determined at a resolution of 2.6 Å. Analysis of the complex shows that the reconstituted GFP is attached to the target protein (sfCherry) in a structurally ordered way. This work opens the way to rapidly creating crystallization variants by reconstituting a target protein bearing the GFP(10-11) hairpin with a variety of GFP(1-9) mutants engineered for favorable crystallization.
Organizational Affiliation:
Bioscience Division, Los Alamos National Laboratory, MS M888, Los Alamos, NM 87545, USA.