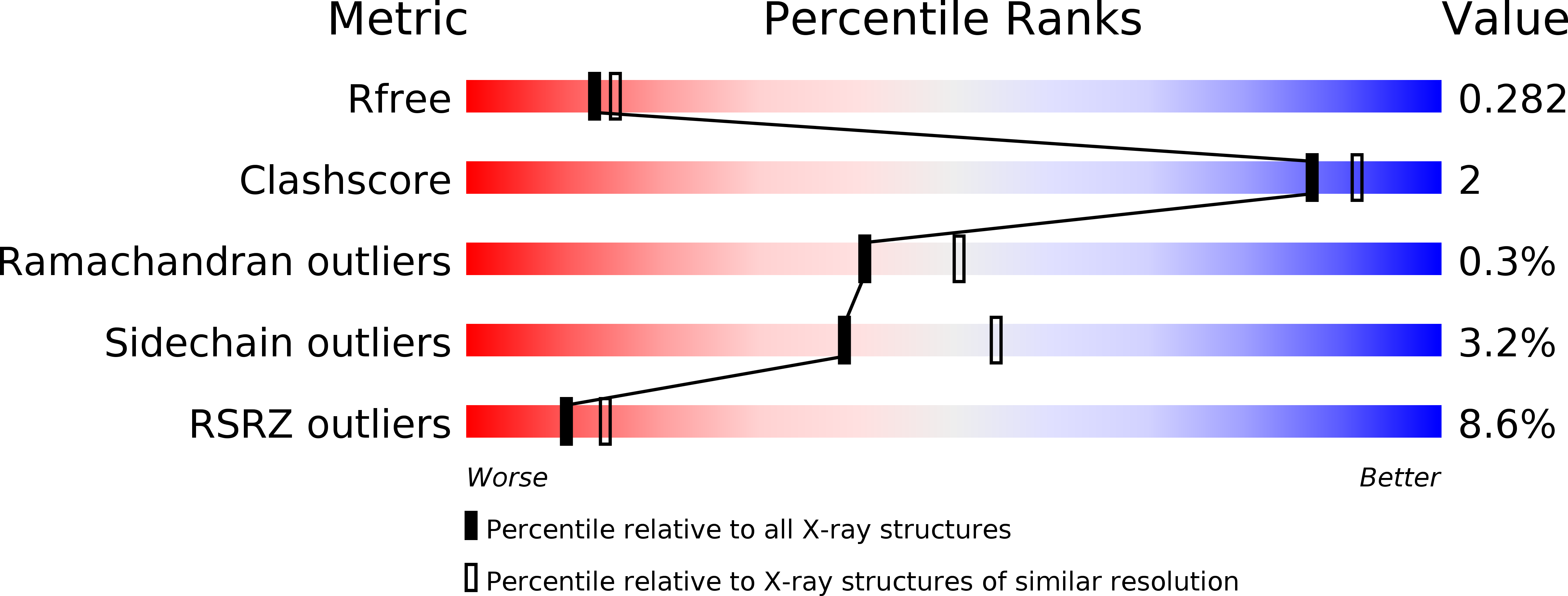
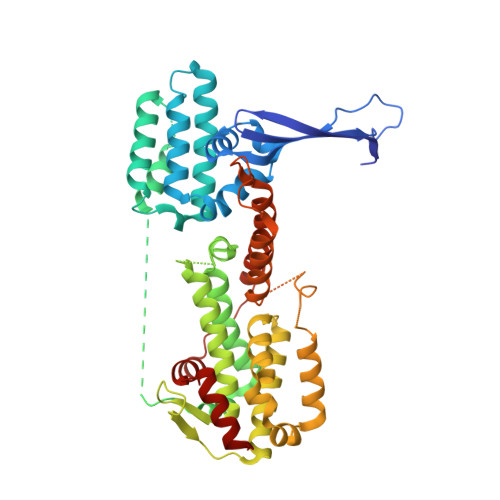
The guanine nucleotide exchange factor Rlf interacts with SH3 domain-containing proteins via a binding site with a preselected conformation.
Popovic, M., Jakobi, A.J., Rensen-de Leeuw, M., Rehmann, H.(2013) J Struct Biol 183: 312-319
- PubMed: 23891840
- DOI: https://doi.org/10.1016/j.jsb.2013.07.009
- Primary Citation of Related Structures:
4JGW - PubMed Abstract:
Rlf is a guanine nucleotide exchange factor for the small G-proteins RalA and RalB and couples Ras- to Ral-signalling. Here the crystal structure of the catalytic module of Rlf consisting of a REM- and a CDC25-homology domain is determined. The structure is distinguished by an extended three stranded β-sheet called the flagpole. The flagpole is a conserved element in the RalGDS family of guanine nucleotide exchange factors and stabilises the orientation of the REM-domain relative to the CDC25-homology domain. A proline-rich sequence in the flagpole is unique to Rlf and several proteins that interact with this sequence by SH3 domains are identified. Conformational pre-selection results in a gain of affinity and contributes to the establishment of SH3 domain selectivity.
Organizational Affiliation:
Department of Molecular Cancer Research, Centre of Biomedical Genetics and Cancer Genomics Centre, University Medical Center Utrecht, Utrecht, The Netherlands.