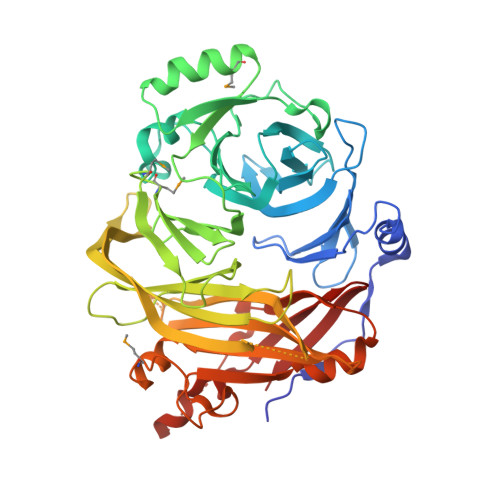
Structural and Functional Studies of the 252 kDa Nucleoporin ELYS Reveal Distinct Roles for Its Three Tethered Domains.
Bilokapic, S., Schwartz, T.U.(2013) Structure 21: 572-580
- PubMed: 23499022
- DOI: https://doi.org/10.1016/j.str.2013.02.006
- Primary Citation of Related Structures:
4I0O - PubMed Abstract:
In metazoa, the nuclear envelope (NE), together with the embedded nuclear pore complexes (NPCs), breaks down and reassembles during cell division. It is suggested that ELYS, a nucleoporin, binds to chromatin in an initial step of postmitotic NPC assembly and subsequently recruits the essential Y-subcomplex, the major scaffolding unit of the NPC. Here, we show that ELYS contains three domains: an N-terminal β-propeller domain, a central α-helical domain, and a C-terminal disordered region. While the disordered region is responsible for the interactions with chromatin, the two preceding domains synergistically mediate tethering to the NPC. We present the crystal structure of the seven-bladed β-propeller domain at 1.9 Å resolution. Analysis of the β-propeller surface reveals the regions that are required for NPC anchorage. We discuss the possible roles of ELYS in the context of the NPC scaffold architecture.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.