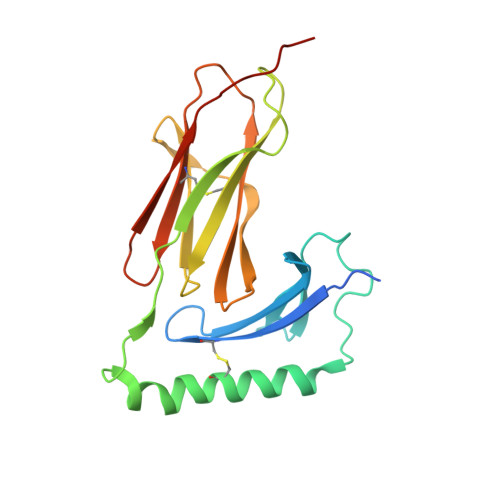
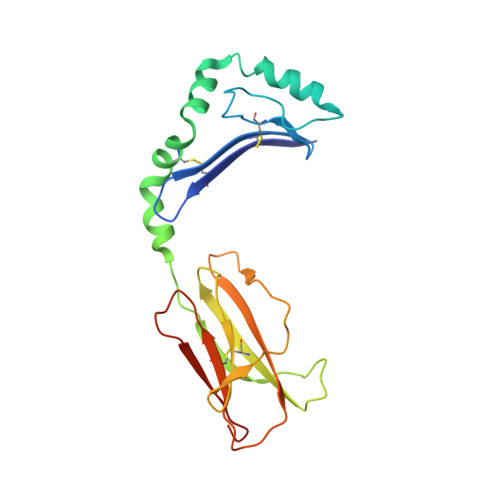
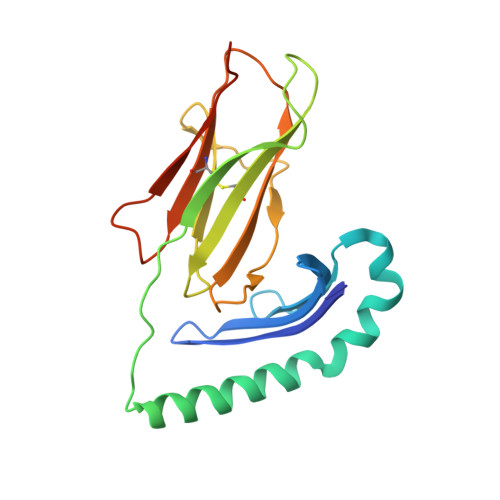
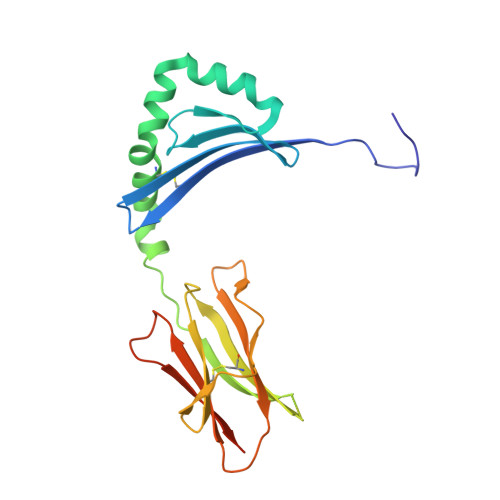
Crystal Structure of the HLA-DM-HLA-DR1 Complex Defines Mechanisms for Rapid Peptide Selection.
Pos, W., Sethi, D.K., Call, M.J., Schulze, M.S., Anders, A.K., Pyrdol, J., Wucherpfennig, K.W.(2012) Cell 151: 1557-1568
- PubMed: 23260142
- DOI: https://doi.org/10.1016/j.cell.2012.11.025
- Primary Citation of Related Structures:
4FQX, 4GBX - PubMed Abstract:
HLA-DR molecules bind microbial peptides in an endosomal compartment and present them on the cell surface for CD4 T cell surveillance. HLA-DM plays a critical role in the endosomal peptide selection process. The structure of the HLA-DM-HLA-DR complex shows major rearrangements of the HLA-DR peptide-binding groove. Flipping of a tryptophan away from the HLA-DR1 P1 pocket enables major conformational changes that position hydrophobic HLA-DR residues into the P1 pocket. These conformational changes accelerate peptide dissociation and stabilize the empty HLA-DR peptide-binding groove. Initially, incoming peptides have access to only part of the HLA-DR groove and need to compete with HLA-DR residues for access to the P2 site and the hydrophobic P1 pocket. This energetic barrier creates a rapid and stringent selection process for the highest-affinity binders. Insertion of peptide residues into the P2 and P1 sites reverses the conformational changes, terminating selection through DM dissociation.
Organizational Affiliation:
Department of Cancer Immunology & AIDS, Dana-Farber Cancer Institute, Boston, MA 02115, USA.