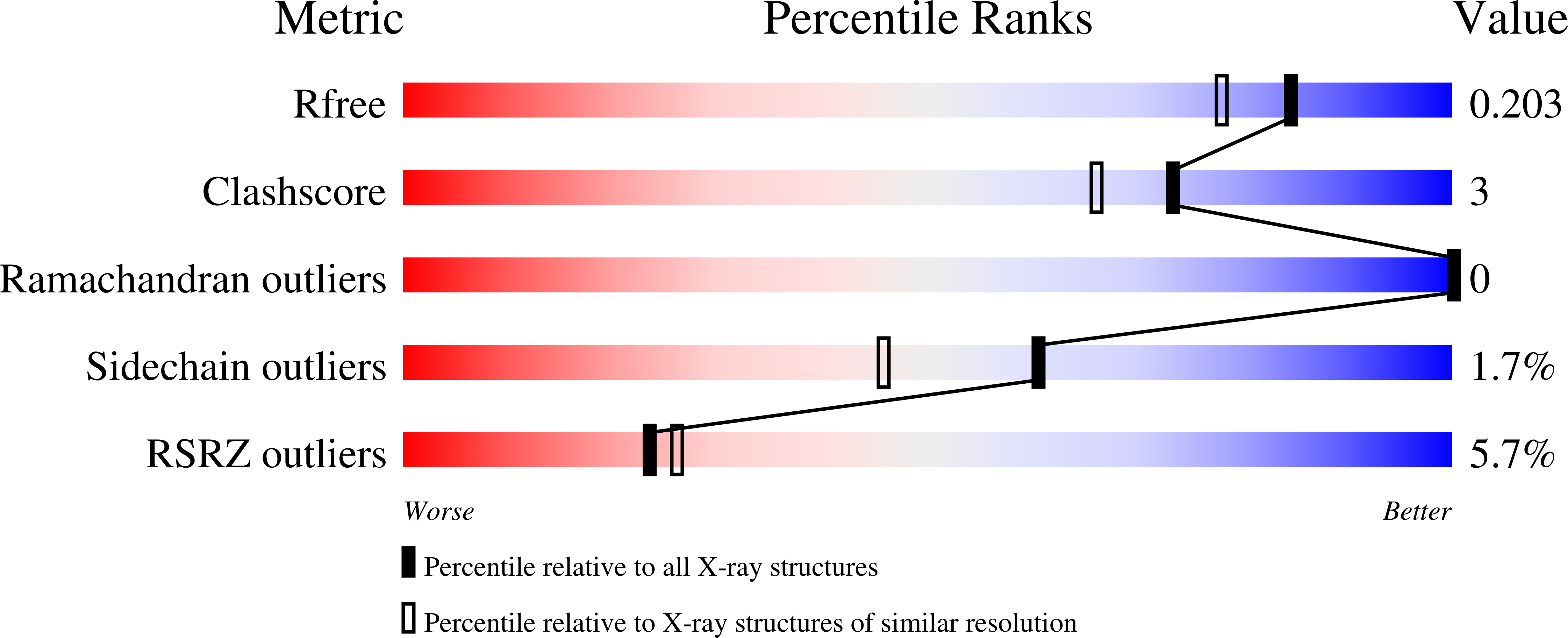
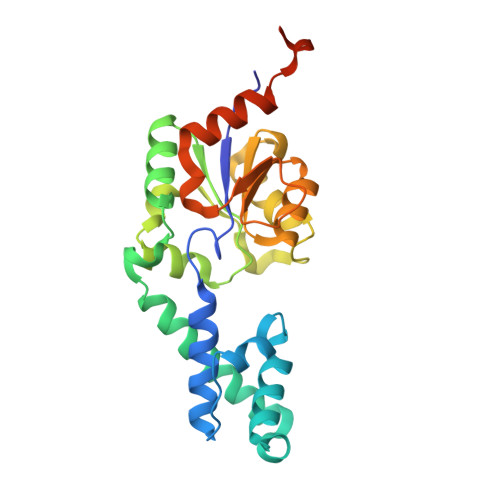
Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
Vetting, M.W., Toro, R., Bhosle, R., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Washington, E., Scott Glenn, A., Chowdhury, S., Evans, B., Hammonds, J., Hillerich, B., Love, J., Seidel, R.D., Imker, H.J., Dunaway-Mariano, D., Allen, K.N., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.